- Privacy Policy

Home » Research Methodology – Types, Examples and writing Guide

Research Methodology – Types, Examples and writing Guide
Table of Contents

Research Methodology
Definition:
Research Methodology refers to the systematic and scientific approach used to conduct research, investigate problems, and gather data and information for a specific purpose. It involves the techniques and procedures used to identify, collect , analyze , and interpret data to answer research questions or solve research problems . Moreover, They are philosophical and theoretical frameworks that guide the research process.
Structure of Research Methodology
Research methodology formats can vary depending on the specific requirements of the research project, but the following is a basic example of a structure for a research methodology section:
I. Introduction
- Provide an overview of the research problem and the need for a research methodology section
- Outline the main research questions and objectives
II. Research Design
- Explain the research design chosen and why it is appropriate for the research question(s) and objectives
- Discuss any alternative research designs considered and why they were not chosen
- Describe the research setting and participants (if applicable)
III. Data Collection Methods
- Describe the methods used to collect data (e.g., surveys, interviews, observations)
- Explain how the data collection methods were chosen and why they are appropriate for the research question(s) and objectives
- Detail any procedures or instruments used for data collection
IV. Data Analysis Methods
- Describe the methods used to analyze the data (e.g., statistical analysis, content analysis )
- Explain how the data analysis methods were chosen and why they are appropriate for the research question(s) and objectives
- Detail any procedures or software used for data analysis
V. Ethical Considerations
- Discuss any ethical issues that may arise from the research and how they were addressed
- Explain how informed consent was obtained (if applicable)
- Detail any measures taken to ensure confidentiality and anonymity
VI. Limitations
- Identify any potential limitations of the research methodology and how they may impact the results and conclusions
VII. Conclusion
- Summarize the key aspects of the research methodology section
- Explain how the research methodology addresses the research question(s) and objectives
Research Methodology Types
Types of Research Methodology are as follows:
Quantitative Research Methodology
This is a research methodology that involves the collection and analysis of numerical data using statistical methods. This type of research is often used to study cause-and-effect relationships and to make predictions.
Qualitative Research Methodology
This is a research methodology that involves the collection and analysis of non-numerical data such as words, images, and observations. This type of research is often used to explore complex phenomena, to gain an in-depth understanding of a particular topic, and to generate hypotheses.
Mixed-Methods Research Methodology
This is a research methodology that combines elements of both quantitative and qualitative research. This approach can be particularly useful for studies that aim to explore complex phenomena and to provide a more comprehensive understanding of a particular topic.
Case Study Research Methodology
This is a research methodology that involves in-depth examination of a single case or a small number of cases. Case studies are often used in psychology, sociology, and anthropology to gain a detailed understanding of a particular individual or group.
Action Research Methodology
This is a research methodology that involves a collaborative process between researchers and practitioners to identify and solve real-world problems. Action research is often used in education, healthcare, and social work.
Experimental Research Methodology
This is a research methodology that involves the manipulation of one or more independent variables to observe their effects on a dependent variable. Experimental research is often used to study cause-and-effect relationships and to make predictions.
Survey Research Methodology
This is a research methodology that involves the collection of data from a sample of individuals using questionnaires or interviews. Survey research is often used to study attitudes, opinions, and behaviors.
Grounded Theory Research Methodology
This is a research methodology that involves the development of theories based on the data collected during the research process. Grounded theory is often used in sociology and anthropology to generate theories about social phenomena.
Research Methodology Example
An Example of Research Methodology could be the following:
Research Methodology for Investigating the Effectiveness of Cognitive Behavioral Therapy in Reducing Symptoms of Depression in Adults
Introduction:
The aim of this research is to investigate the effectiveness of cognitive-behavioral therapy (CBT) in reducing symptoms of depression in adults. To achieve this objective, a randomized controlled trial (RCT) will be conducted using a mixed-methods approach.
Research Design:
The study will follow a pre-test and post-test design with two groups: an experimental group receiving CBT and a control group receiving no intervention. The study will also include a qualitative component, in which semi-structured interviews will be conducted with a subset of participants to explore their experiences of receiving CBT.
Participants:
Participants will be recruited from community mental health clinics in the local area. The sample will consist of 100 adults aged 18-65 years old who meet the diagnostic criteria for major depressive disorder. Participants will be randomly assigned to either the experimental group or the control group.
Intervention :
The experimental group will receive 12 weekly sessions of CBT, each lasting 60 minutes. The intervention will be delivered by licensed mental health professionals who have been trained in CBT. The control group will receive no intervention during the study period.
Data Collection:
Quantitative data will be collected through the use of standardized measures such as the Beck Depression Inventory-II (BDI-II) and the Generalized Anxiety Disorder-7 (GAD-7). Data will be collected at baseline, immediately after the intervention, and at a 3-month follow-up. Qualitative data will be collected through semi-structured interviews with a subset of participants from the experimental group. The interviews will be conducted at the end of the intervention period, and will explore participants’ experiences of receiving CBT.
Data Analysis:
Quantitative data will be analyzed using descriptive statistics, t-tests, and mixed-model analyses of variance (ANOVA) to assess the effectiveness of the intervention. Qualitative data will be analyzed using thematic analysis to identify common themes and patterns in participants’ experiences of receiving CBT.
Ethical Considerations:
This study will comply with ethical guidelines for research involving human subjects. Participants will provide informed consent before participating in the study, and their privacy and confidentiality will be protected throughout the study. Any adverse events or reactions will be reported and managed appropriately.
Data Management:
All data collected will be kept confidential and stored securely using password-protected databases. Identifying information will be removed from qualitative data transcripts to ensure participants’ anonymity.
Limitations:
One potential limitation of this study is that it only focuses on one type of psychotherapy, CBT, and may not generalize to other types of therapy or interventions. Another limitation is that the study will only include participants from community mental health clinics, which may not be representative of the general population.
Conclusion:
This research aims to investigate the effectiveness of CBT in reducing symptoms of depression in adults. By using a randomized controlled trial and a mixed-methods approach, the study will provide valuable insights into the mechanisms underlying the relationship between CBT and depression. The results of this study will have important implications for the development of effective treatments for depression in clinical settings.
How to Write Research Methodology
Writing a research methodology involves explaining the methods and techniques you used to conduct research, collect data, and analyze results. It’s an essential section of any research paper or thesis, as it helps readers understand the validity and reliability of your findings. Here are the steps to write a research methodology:
- Start by explaining your research question: Begin the methodology section by restating your research question and explaining why it’s important. This helps readers understand the purpose of your research and the rationale behind your methods.
- Describe your research design: Explain the overall approach you used to conduct research. This could be a qualitative or quantitative research design, experimental or non-experimental, case study or survey, etc. Discuss the advantages and limitations of the chosen design.
- Discuss your sample: Describe the participants or subjects you included in your study. Include details such as their demographics, sampling method, sample size, and any exclusion criteria used.
- Describe your data collection methods : Explain how you collected data from your participants. This could include surveys, interviews, observations, questionnaires, or experiments. Include details on how you obtained informed consent, how you administered the tools, and how you minimized the risk of bias.
- Explain your data analysis techniques: Describe the methods you used to analyze the data you collected. This could include statistical analysis, content analysis, thematic analysis, or discourse analysis. Explain how you dealt with missing data, outliers, and any other issues that arose during the analysis.
- Discuss the validity and reliability of your research : Explain how you ensured the validity and reliability of your study. This could include measures such as triangulation, member checking, peer review, or inter-coder reliability.
- Acknowledge any limitations of your research: Discuss any limitations of your study, including any potential threats to validity or generalizability. This helps readers understand the scope of your findings and how they might apply to other contexts.
- Provide a summary: End the methodology section by summarizing the methods and techniques you used to conduct your research. This provides a clear overview of your research methodology and helps readers understand the process you followed to arrive at your findings.
When to Write Research Methodology
Research methodology is typically written after the research proposal has been approved and before the actual research is conducted. It should be written prior to data collection and analysis, as it provides a clear roadmap for the research project.
The research methodology is an important section of any research paper or thesis, as it describes the methods and procedures that will be used to conduct the research. It should include details about the research design, data collection methods, data analysis techniques, and any ethical considerations.
The methodology should be written in a clear and concise manner, and it should be based on established research practices and standards. It is important to provide enough detail so that the reader can understand how the research was conducted and evaluate the validity of the results.
Applications of Research Methodology
Here are some of the applications of research methodology:
- To identify the research problem: Research methodology is used to identify the research problem, which is the first step in conducting any research.
- To design the research: Research methodology helps in designing the research by selecting the appropriate research method, research design, and sampling technique.
- To collect data: Research methodology provides a systematic approach to collect data from primary and secondary sources.
- To analyze data: Research methodology helps in analyzing the collected data using various statistical and non-statistical techniques.
- To test hypotheses: Research methodology provides a framework for testing hypotheses and drawing conclusions based on the analysis of data.
- To generalize findings: Research methodology helps in generalizing the findings of the research to the target population.
- To develop theories : Research methodology is used to develop new theories and modify existing theories based on the findings of the research.
- To evaluate programs and policies : Research methodology is used to evaluate the effectiveness of programs and policies by collecting data and analyzing it.
- To improve decision-making: Research methodology helps in making informed decisions by providing reliable and valid data.
Purpose of Research Methodology
Research methodology serves several important purposes, including:
- To guide the research process: Research methodology provides a systematic framework for conducting research. It helps researchers to plan their research, define their research questions, and select appropriate methods and techniques for collecting and analyzing data.
- To ensure research quality: Research methodology helps researchers to ensure that their research is rigorous, reliable, and valid. It provides guidelines for minimizing bias and error in data collection and analysis, and for ensuring that research findings are accurate and trustworthy.
- To replicate research: Research methodology provides a clear and detailed account of the research process, making it possible for other researchers to replicate the study and verify its findings.
- To advance knowledge: Research methodology enables researchers to generate new knowledge and to contribute to the body of knowledge in their field. It provides a means for testing hypotheses, exploring new ideas, and discovering new insights.
- To inform decision-making: Research methodology provides evidence-based information that can inform policy and decision-making in a variety of fields, including medicine, public health, education, and business.
Advantages of Research Methodology
Research methodology has several advantages that make it a valuable tool for conducting research in various fields. Here are some of the key advantages of research methodology:
- Systematic and structured approach : Research methodology provides a systematic and structured approach to conducting research, which ensures that the research is conducted in a rigorous and comprehensive manner.
- Objectivity : Research methodology aims to ensure objectivity in the research process, which means that the research findings are based on evidence and not influenced by personal bias or subjective opinions.
- Replicability : Research methodology ensures that research can be replicated by other researchers, which is essential for validating research findings and ensuring their accuracy.
- Reliability : Research methodology aims to ensure that the research findings are reliable, which means that they are consistent and can be depended upon.
- Validity : Research methodology ensures that the research findings are valid, which means that they accurately reflect the research question or hypothesis being tested.
- Efficiency : Research methodology provides a structured and efficient way of conducting research, which helps to save time and resources.
- Flexibility : Research methodology allows researchers to choose the most appropriate research methods and techniques based on the research question, data availability, and other relevant factors.
- Scope for innovation: Research methodology provides scope for innovation and creativity in designing research studies and developing new research techniques.
Research Methodology Vs Research Methods
| Research Methodology | Research Methods |
|---|---|
| Research methodology refers to the philosophical and theoretical frameworks that guide the research process. | refer to the techniques and procedures used to collect and analyze data. |
| It is concerned with the underlying principles and assumptions of research. | It is concerned with the practical aspects of research. |
| It provides a rationale for why certain research methods are used. | It determines the specific steps that will be taken to conduct research. |
| It is broader in scope and involves understanding the overall approach to research. | It is narrower in scope and focuses on specific techniques and tools used in research. |
| It is concerned with identifying research questions, defining the research problem, and formulating hypotheses. | It is concerned with collecting data, analyzing data, and interpreting results. |
| It is concerned with the validity and reliability of research. | It is concerned with the accuracy and precision of data. |
| It is concerned with the ethical considerations of research. | It is concerned with the practical considerations of research. |
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

Research Methods – Types, Examples and Guide

Scope of the Research – Writing Guide and...

References in Research – Types, Examples and...

Research Summary – Structure, Examples and...

Research Paper Format – Types, Examples and...

Ethical Considerations – Types, Examples and...
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, generate accurate citations for free.
- Knowledge Base
Methodology
Research Methods | Definitions, Types, Examples
Research methods are specific procedures for collecting and analyzing data. Developing your research methods is an integral part of your research design . When planning your methods, there are two key decisions you will make.
First, decide how you will collect data . Your methods depend on what type of data you need to answer your research question :
- Qualitative vs. quantitative : Will your data take the form of words or numbers?
- Primary vs. secondary : Will you collect original data yourself, or will you use data that has already been collected by someone else?
- Descriptive vs. experimental : Will you take measurements of something as it is, or will you perform an experiment?
Second, decide how you will analyze the data .
- For quantitative data, you can use statistical analysis methods to test relationships between variables.
- For qualitative data, you can use methods such as thematic analysis to interpret patterns and meanings in the data.
Table of contents
Methods for collecting data, examples of data collection methods, methods for analyzing data, examples of data analysis methods, other interesting articles, frequently asked questions about research methods.
Data is the information that you collect for the purposes of answering your research question . The type of data you need depends on the aims of your research.
Qualitative vs. quantitative data
Your choice of qualitative or quantitative data collection depends on the type of knowledge you want to develop.
For questions about ideas, experiences and meanings, or to study something that can’t be described numerically, collect qualitative data .
If you want to develop a more mechanistic understanding of a topic, or your research involves hypothesis testing , collect quantitative data .
| Qualitative | to broader populations. . | |
|---|---|---|
| Quantitative | . |
You can also take a mixed methods approach , where you use both qualitative and quantitative research methods.
Primary vs. secondary research
Primary research is any original data that you collect yourself for the purposes of answering your research question (e.g. through surveys , observations and experiments ). Secondary research is data that has already been collected by other researchers (e.g. in a government census or previous scientific studies).
If you are exploring a novel research question, you’ll probably need to collect primary data . But if you want to synthesize existing knowledge, analyze historical trends, or identify patterns on a large scale, secondary data might be a better choice.
| Primary | . | methods. |
|---|---|---|
| Secondary |
Descriptive vs. experimental data
In descriptive research , you collect data about your study subject without intervening. The validity of your research will depend on your sampling method .
In experimental research , you systematically intervene in a process and measure the outcome. The validity of your research will depend on your experimental design .
To conduct an experiment, you need to be able to vary your independent variable , precisely measure your dependent variable, and control for confounding variables . If it’s practically and ethically possible, this method is the best choice for answering questions about cause and effect.
| Descriptive | . . | |
|---|---|---|
| Experimental |
Here's why students love Scribbr's proofreading services
Discover proofreading & editing
| Research method | Primary or secondary? | Qualitative or quantitative? | When to use |
|---|---|---|---|
| Primary | Quantitative | To test cause-and-effect relationships. | |
| Primary | Quantitative | To understand general characteristics of a population. | |
| Interview/focus group | Primary | Qualitative | To gain more in-depth understanding of a topic. |
| Observation | Primary | Either | To understand how something occurs in its natural setting. |
| Secondary | Either | To situate your research in an existing body of work, or to evaluate trends within a research topic. | |
| Either | Either | To gain an in-depth understanding of a specific group or context, or when you don’t have the resources for a large study. |
Your data analysis methods will depend on the type of data you collect and how you prepare it for analysis.
Data can often be analyzed both quantitatively and qualitatively. For example, survey responses could be analyzed qualitatively by studying the meanings of responses or quantitatively by studying the frequencies of responses.
Qualitative analysis methods
Qualitative analysis is used to understand words, ideas, and experiences. You can use it to interpret data that was collected:
- From open-ended surveys and interviews , literature reviews , case studies , ethnographies , and other sources that use text rather than numbers.
- Using non-probability sampling methods .
Qualitative analysis tends to be quite flexible and relies on the researcher’s judgement, so you have to reflect carefully on your choices and assumptions and be careful to avoid research bias .
Quantitative analysis methods
Quantitative analysis uses numbers and statistics to understand frequencies, averages and correlations (in descriptive studies) or cause-and-effect relationships (in experiments).
You can use quantitative analysis to interpret data that was collected either:
- During an experiment .
- Using probability sampling methods .
Because the data is collected and analyzed in a statistically valid way, the results of quantitative analysis can be easily standardized and shared among researchers.
| Research method | Qualitative or quantitative? | When to use |
|---|---|---|
| Quantitative | To analyze data collected in a statistically valid manner (e.g. from experiments, surveys, and observations). | |
| Meta-analysis | Quantitative | To statistically analyze the results of a large collection of studies. Can only be applied to studies that collected data in a statistically valid manner. |
| Qualitative | To analyze data collected from interviews, , or textual sources. To understand general themes in the data and how they are communicated. | |
| Either | To analyze large volumes of textual or visual data collected from surveys, literature reviews, or other sources. Can be quantitative (i.e. frequencies of words) or qualitative (i.e. meanings of words). |
Prevent plagiarism. Run a free check.
If you want to know more about statistics , methodology , or research bias , make sure to check out some of our other articles with explanations and examples.
- Chi square test of independence
- Statistical power
- Descriptive statistics
- Degrees of freedom
- Pearson correlation
- Null hypothesis
- Double-blind study
- Case-control study
- Research ethics
- Data collection
- Hypothesis testing
- Structured interviews
Research bias
- Hawthorne effect
- Unconscious bias
- Recall bias
- Halo effect
- Self-serving bias
- Information bias
Quantitative research deals with numbers and statistics, while qualitative research deals with words and meanings.
Quantitative methods allow you to systematically measure variables and test hypotheses . Qualitative methods allow you to explore concepts and experiences in more detail.
In mixed methods research , you use both qualitative and quantitative data collection and analysis methods to answer your research question .
A sample is a subset of individuals from a larger population . Sampling means selecting the group that you will actually collect data from in your research. For example, if you are researching the opinions of students in your university, you could survey a sample of 100 students.
In statistics, sampling allows you to test a hypothesis about the characteristics of a population.
The research methods you use depend on the type of data you need to answer your research question .
- If you want to measure something or test a hypothesis , use quantitative methods . If you want to explore ideas, thoughts and meanings, use qualitative methods .
- If you want to analyze a large amount of readily-available data, use secondary data. If you want data specific to your purposes with control over how it is generated, collect primary data.
- If you want to establish cause-and-effect relationships between variables , use experimental methods. If you want to understand the characteristics of a research subject, use descriptive methods.
Methodology refers to the overarching strategy and rationale of your research project . It involves studying the methods used in your field and the theories or principles behind them, in order to develop an approach that matches your objectives.
Methods are the specific tools and procedures you use to collect and analyze data (for example, experiments, surveys , and statistical tests ).
In shorter scientific papers, where the aim is to report the findings of a specific study, you might simply describe what you did in a methods section .
In a longer or more complex research project, such as a thesis or dissertation , you will probably include a methodology section , where you explain your approach to answering the research questions and cite relevant sources to support your choice of methods.
Is this article helpful?
Other students also liked, writing strong research questions | criteria & examples.
- What Is a Research Design | Types, Guide & Examples
- Data Collection | Definition, Methods & Examples
More interesting articles
- Between-Subjects Design | Examples, Pros, & Cons
- Cluster Sampling | A Simple Step-by-Step Guide with Examples
- Confounding Variables | Definition, Examples & Controls
- Construct Validity | Definition, Types, & Examples
- Content Analysis | Guide, Methods & Examples
- Control Groups and Treatment Groups | Uses & Examples
- Control Variables | What Are They & Why Do They Matter?
- Correlation vs. Causation | Difference, Designs & Examples
- Correlational Research | When & How to Use
- Critical Discourse Analysis | Definition, Guide & Examples
- Cross-Sectional Study | Definition, Uses & Examples
- Descriptive Research | Definition, Types, Methods & Examples
- Ethical Considerations in Research | Types & Examples
- Explanatory and Response Variables | Definitions & Examples
- Explanatory Research | Definition, Guide, & Examples
- Exploratory Research | Definition, Guide, & Examples
- External Validity | Definition, Types, Threats & Examples
- Extraneous Variables | Examples, Types & Controls
- Guide to Experimental Design | Overview, Steps, & Examples
- How Do You Incorporate an Interview into a Dissertation? | Tips
- How to Do Thematic Analysis | Step-by-Step Guide & Examples
- How to Write a Literature Review | Guide, Examples, & Templates
- How to Write a Strong Hypothesis | Steps & Examples
- Inclusion and Exclusion Criteria | Examples & Definition
- Independent vs. Dependent Variables | Definition & Examples
- Inductive Reasoning | Types, Examples, Explanation
- Inductive vs. Deductive Research Approach | Steps & Examples
- Internal Validity in Research | Definition, Threats, & Examples
- Internal vs. External Validity | Understanding Differences & Threats
- Longitudinal Study | Definition, Approaches & Examples
- Mediator vs. Moderator Variables | Differences & Examples
- Mixed Methods Research | Definition, Guide & Examples
- Multistage Sampling | Introductory Guide & Examples
- Naturalistic Observation | Definition, Guide & Examples
- Operationalization | A Guide with Examples, Pros & Cons
- Population vs. Sample | Definitions, Differences & Examples
- Primary Research | Definition, Types, & Examples
- Qualitative vs. Quantitative Research | Differences, Examples & Methods
- Quasi-Experimental Design | Definition, Types & Examples
- Questionnaire Design | Methods, Question Types & Examples
- Random Assignment in Experiments | Introduction & Examples
- Random vs. Systematic Error | Definition & Examples
- Reliability vs. Validity in Research | Difference, Types and Examples
- Reproducibility vs Replicability | Difference & Examples
- Reproducibility vs. Replicability | Difference & Examples
- Sampling Methods | Types, Techniques & Examples
- Semi-Structured Interview | Definition, Guide & Examples
- Simple Random Sampling | Definition, Steps & Examples
- Single, Double, & Triple Blind Study | Definition & Examples
- Stratified Sampling | Definition, Guide & Examples
- Structured Interview | Definition, Guide & Examples
- Survey Research | Definition, Examples & Methods
- Systematic Review | Definition, Example, & Guide
- Systematic Sampling | A Step-by-Step Guide with Examples
- Textual Analysis | Guide, 3 Approaches & Examples
- The 4 Types of Reliability in Research | Definitions & Examples
- The 4 Types of Validity in Research | Definitions & Examples
- Transcribing an Interview | 5 Steps & Transcription Software
- Triangulation in Research | Guide, Types, Examples
- Types of Interviews in Research | Guide & Examples
- Types of Research Designs Compared | Guide & Examples
- Types of Variables in Research & Statistics | Examples
- Unstructured Interview | Definition, Guide & Examples
- What Is a Case Study? | Definition, Examples & Methods
- What Is a Case-Control Study? | Definition & Examples
- What Is a Cohort Study? | Definition & Examples
- What Is a Conceptual Framework? | Tips & Examples
- What Is a Controlled Experiment? | Definitions & Examples
- What Is a Double-Barreled Question?
- What Is a Focus Group? | Step-by-Step Guide & Examples
- What Is a Likert Scale? | Guide & Examples
- What Is a Prospective Cohort Study? | Definition & Examples
- What Is a Retrospective Cohort Study? | Definition & Examples
- What Is Action Research? | Definition & Examples
- What Is an Observational Study? | Guide & Examples
- What Is Concurrent Validity? | Definition & Examples
- What Is Content Validity? | Definition & Examples
- What Is Convenience Sampling? | Definition & Examples
- What Is Convergent Validity? | Definition & Examples
- What Is Criterion Validity? | Definition & Examples
- What Is Data Cleansing? | Definition, Guide & Examples
- What Is Deductive Reasoning? | Explanation & Examples
- What Is Discriminant Validity? | Definition & Example
- What Is Ecological Validity? | Definition & Examples
- What Is Ethnography? | Definition, Guide & Examples
- What Is Face Validity? | Guide, Definition & Examples
- What Is Non-Probability Sampling? | Types & Examples
- What Is Participant Observation? | Definition & Examples
- What Is Peer Review? | Types & Examples
- What Is Predictive Validity? | Examples & Definition
- What Is Probability Sampling? | Types & Examples
- What Is Purposive Sampling? | Definition & Examples
- What Is Qualitative Observation? | Definition & Examples
- What Is Qualitative Research? | Methods & Examples
- What Is Quantitative Observation? | Definition & Examples
- What Is Quantitative Research? | Definition, Uses & Methods
Get unlimited documents corrected
✔ Free APA citation check included ✔ Unlimited document corrections ✔ Specialized in correcting academic texts

What Is Research Methodology?

I f you’re new to formal academic research, it’s quite likely that you’re feeling a little overwhelmed by all the technical lingo that gets thrown around. And who could blame you – “research methodology”, “research methods”, “sampling strategies”… it all seems never-ending!
In this post, we’ll demystify the landscape with plain-language explanations and loads of examples (including easy-to-follow videos), so that you can approach your dissertation, thesis or research project with confidence. Let’s get started.
Research Methodology 101
- What exactly research methodology means
- What qualitative , quantitative and mixed methods are
- What sampling strategy is
- What data collection methods are
- What data analysis methods are
- How to choose your research methodology
- Example of a research methodology

What is research methodology?
Research methodology simply refers to the practical “how” of a research study. More specifically, it’s about how a researcher systematically designs a study to ensure valid and reliable results that address the research aims, objectives and research questions . Specifically, how the researcher went about deciding:
- What type of data to collect (e.g., qualitative or quantitative data )
- Who to collect it from (i.e., the sampling strategy )
- How to collect it (i.e., the data collection method )
- How to analyse it (i.e., the data analysis methods )
Within any formal piece of academic research (be it a dissertation, thesis or journal article), you’ll find a research methodology chapter or section which covers the aspects mentioned above. Importantly, a good methodology chapter explains not just what methodological choices were made, but also explains why they were made. In other words, the methodology chapter should justify the design choices, by showing that the chosen methods and techniques are the best fit for the research aims, objectives and research questions.
So, it’s the same as research design?
Not quite. As we mentioned, research methodology refers to the collection of practical decisions regarding what data you’ll collect, from who, how you’ll collect it and how you’ll analyse it. Research design, on the other hand, is more about the overall strategy you’ll adopt in your study. For example, whether you’ll use an experimental design in which you manipulate one variable while controlling others. You can learn more about research design and the various design types here .
Need a helping hand?
What are qualitative, quantitative and mixed-methods?
Qualitative, quantitative and mixed-methods are different types of methodological approaches, distinguished by their focus on words , numbers or both . This is a bit of an oversimplification, but its a good starting point for understanding.
Let’s take a closer look.
Qualitative research refers to research which focuses on collecting and analysing words (written or spoken) and textual or visual data, whereas quantitative research focuses on measurement and testing using numerical data . Qualitative analysis can also focus on other “softer” data points, such as body language or visual elements.
It’s quite common for a qualitative methodology to be used when the research aims and research questions are exploratory in nature. For example, a qualitative methodology might be used to understand peoples’ perceptions about an event that took place, or a political candidate running for president.
Contrasted to this, a quantitative methodology is typically used when the research aims and research questions are confirmatory in nature. For example, a quantitative methodology might be used to measure the relationship between two variables (e.g. personality type and likelihood to commit a crime) or to test a set of hypotheses .
As you’ve probably guessed, the mixed-method methodology attempts to combine the best of both qualitative and quantitative methodologies to integrate perspectives and create a rich picture. If you’d like to learn more about these three methodological approaches, be sure to watch our explainer video below.
What is sampling strategy?
Simply put, sampling is about deciding who (or where) you’re going to collect your data from . Why does this matter? Well, generally it’s not possible to collect data from every single person in your group of interest (this is called the “population”), so you’ll need to engage a smaller portion of that group that’s accessible and manageable (this is called the “sample”).
How you go about selecting the sample (i.e., your sampling strategy) will have a major impact on your study. There are many different sampling methods you can choose from, but the two overarching categories are probability sampling and non-probability sampling .
Probability sampling involves using a completely random sample from the group of people you’re interested in. This is comparable to throwing the names all potential participants into a hat, shaking it up, and picking out the “winners”. By using a completely random sample, you’ll minimise the risk of selection bias and the results of your study will be more generalisable to the entire population.
Non-probability sampling , on the other hand, doesn’t use a random sample . For example, it might involve using a convenience sample, which means you’d only interview or survey people that you have access to (perhaps your friends, family or work colleagues), rather than a truly random sample. With non-probability sampling, the results are typically not generalisable .
To learn more about sampling methods, be sure to check out the video below.
What are data collection methods?
As the name suggests, data collection methods simply refers to the way in which you go about collecting the data for your study. Some of the most common data collection methods include:
- Interviews (which can be unstructured, semi-structured or structured)
- Focus groups and group interviews
- Surveys (online or physical surveys)
- Observations (watching and recording activities)
- Biophysical measurements (e.g., blood pressure, heart rate, etc.)
- Documents and records (e.g., financial reports, court records, etc.)
The choice of which data collection method to use depends on your overall research aims and research questions , as well as practicalities and resource constraints. For example, if your research is exploratory in nature, qualitative methods such as interviews and focus groups would likely be a good fit. Conversely, if your research aims to measure specific variables or test hypotheses, large-scale surveys that produce large volumes of numerical data would likely be a better fit.
What are data analysis methods?
Data analysis methods refer to the methods and techniques that you’ll use to make sense of your data. These can be grouped according to whether the research is qualitative (words-based) or quantitative (numbers-based).
Popular data analysis methods in qualitative research include:
- Qualitative content analysis
- Thematic analysis
- Discourse analysis
- Narrative analysis
- Interpretative phenomenological analysis (IPA)
- Visual analysis (of photographs, videos, art, etc.)
Qualitative data analysis all begins with data coding , after which an analysis method is applied. In some cases, more than one analysis method is used, depending on the research aims and research questions . In the video below, we explore some common qualitative analysis methods, along with practical examples.
- Descriptive statistics (e.g. means, medians, modes )
- Inferential statistics (e.g. correlation, regression, structural equation modelling)
How do I choose a research methodology?
As you’ve probably picked up by now, your research aims and objectives have a major influence on the research methodology . So, the starting point for developing your research methodology is to take a step back and look at the big picture of your research, before you make methodology decisions. The first question you need to ask yourself is whether your research is exploratory or confirmatory in nature.
If your research aims and objectives are primarily exploratory in nature, your research will likely be qualitative and therefore you might consider qualitative data collection methods (e.g. interviews) and analysis methods (e.g. qualitative content analysis).
Conversely, if your research aims and objective are looking to measure or test something (i.e. they’re confirmatory), then your research will quite likely be quantitative in nature, and you might consider quantitative data collection methods (e.g. surveys) and analyses (e.g. statistical analysis).
Designing your research and working out your methodology is a large topic, which we cover extensively on the blog . For now, however, the key takeaway is that you should always start with your research aims, objectives and research questions (the golden thread). Every methodological choice you make needs align with those three components.
Example of a research methodology chapter
In the video below, we provide a detailed walkthrough of a research methodology from an actual dissertation, as well as an overview of our free methodology template .

Learn More About Methodology

Triangulation: The Ultimate Credibility Enhancer
Triangulation is one of the best ways to enhance the credibility of your research. Learn about the different options here.

Research Limitations 101: What You Need To Know
Learn everything you need to know about research limitations (AKA limitations of the study). Includes practical examples from real studies.

In Vivo Coding 101: Full Explainer With Examples
Learn about in vivo coding, a popular qualitative coding technique ideal for studies where the nuances of language are central to the aims.

Process Coding 101: Full Explainer With Examples
Learn about process coding, a popular qualitative coding technique ideal for studies exploring processes, actions and changes over time.

Qualitative Coding 101: Inductive, Deductive & Hybrid Coding
Inductive, Deductive & Abductive Coding Qualitative Coding Approaches Explained...
📄 FREE TEMPLATES
Research Topic Ideation
Proposal Writing
Literature Review
Methodology & Analysis
Academic Writing
Referencing & Citing
Apps, Tools & Tricks
The Grad Coach Podcast
199 Comments
Thank you for this simple yet comprehensive and easy to digest presentation. God Bless!
You’re most welcome, Leo. Best of luck with your research!
I found it very useful. many thanks
This is really directional. A make-easy research knowledge.
Thank you for this, I think will help my research proposal
Thanks for good interpretation,well understood.
Good morning sorry I want to the search topic
Thank u more
Thank you, your explanation is simple and very helpful.
Very educative a.nd exciting platform. A bigger thank you and I’ll like to always be with you
That’s the best analysis
So simple yet so insightful. Thank you.
This really easy to read as it is self-explanatory. Very much appreciated…
Thanks for this. It’s so helpful and explicit. For those elements highlighted in orange, they were good sources of referrals for concepts I didn’t understand. A million thanks for this.
Good morning, I have been reading your research lessons through out a period of times. They are important, impressive and clear. Want to subscribe and be and be active with you.
Thankyou So much Sir Derek…
Good morning thanks so much for the on line lectures am a student of university of Makeni.select a research topic and deliberate on it so that we’ll continue to understand more.sorry that’s a suggestion.
Beautiful presentation. I love it.
please provide a research mehodology example for zoology
It’s very educative and well explained
Thanks for the concise and informative data.
This is really good for students to be safe and well understand that research is all about
Thank you so much Derek sir🖤🙏🤗
Very simple and reliable
This is really helpful. Thanks alot. God bless you.
very useful, Thank you very much..
thanks a lot its really useful
in a nutshell..thank you!
Thanks for updating my understanding on this aspect of my Thesis writing.
thank you so much my through this video am competently going to do a good job my thesis
Thanks a lot. Very simple to understand. I appreciate 🙏
Very simple but yet insightful Thank you
This has been an eye opening experience. Thank you grad coach team.
Very useful message for research scholars
Really very helpful thank you
yes you are right and i’m left
Research methodology with a simplest way i have never seen before this article.
wow thank u so much
Good morning thanks so much for the on line lectures am a student of university of Makeni.select a research topic and deliberate on is so that we will continue to understand more.sorry that’s a suggestion.
Very precise and informative.
Thanks for simplifying these terms for us, really appreciate it.
Thanks this has really helped me. It is very easy to understand.
I found the notes and the presentation assisting and opening my understanding on research methodology
Good presentation
Im so glad you clarified my misconceptions. Im now ready to fry my onions. Thank you so much. God bless
Thank you a lot.
thanks for the easy way of learning and desirable presentation.
Thanks a lot. I am inspired
Well written
I am writing a APA Format paper . I using questionnaire with 120 STDs teacher for my participant. Can you write me mthology for this research. Send it through email sent. Just need a sample as an example please. My topic is ” impacts of overcrowding on students learning
Thanks for your comment.
We can’t write your methodology for you. If you’re looking for samples, you should be able to find some sample methodologies on Google. Alternatively, you can download some previous dissertations from a dissertation directory and have a look at the methodology chapters therein.
All the best with your research.
Thank you so much for this!! God Bless
Thank you. Explicit explanation
Thank you, Derek and Kerryn, for making this simple to understand. I’m currently at the inception stage of my research.
Thnks a lot , this was very usefull on my assignment
excellent explanation
I’m currently working on my master’s thesis, thanks for this! I’m certain that I will use Qualitative methodology.
Thanks a lot for this concise piece, it was quite relieving and helpful. God bless you BIG…
I am currently doing my dissertation proposal and I am sure that I will do quantitative research. Thank you very much it was extremely helpful.
Very interesting and informative yet I would like to know about examples of Research Questions as well, if possible.
I’m about to submit a research presentation, I have come to understand from your simplification on understanding research methodology. My research will be mixed methodology, qualitative as well as quantitative. So aim and objective of mixed method would be both exploratory and confirmatory. Thanks you very much for your guidance.
OMG thanks for that, you’re a life saver. You covered all the points I needed. Thank you so much ❤️ ❤️ ❤️
Thank you immensely for this simple, easy to comprehend explanation of data collection methods. I have been stuck here for months 😩. Glad I found your piece. Super insightful.
I’m going to write synopsis which will be quantitative research method and I don’t know how to frame my topic, can I kindly get some ideas..
Thanks for this, I was really struggling.
This was really informative I was struggling but this helped me.
Thanks a lot for this information, simple and straightforward. I’m a last year student from the University of South Africa UNISA South Africa.
its very much informative and understandable. I have enlightened.
An interesting nice exploration of a topic.
Thank you. Accurate and simple🥰
This article was really helpful, it helped me understanding the basic concepts of the topic Research Methodology. The examples were very clear, and easy to understand. I would like to visit this website again. Thank you so much for such a great explanation of the subject.
Thanks dude
Thank you Doctor Derek for this wonderful piece, please help to provide your details for reference purpose. God bless.
Many compliments to you
Great work , thank you very much for the simple explanation
Thank you. I had to give a presentation on this topic. I have looked everywhere on the internet but this is the best and simple explanation.
thank you, its very informative.
Well explained. Now I know my research methodology will be qualitative and exploratory. Thank you so much, keep up the good work
Well explained, thank you very much.
This is good explanation, I have understood the different methods of research. Thanks a lot.
Great work…very well explanation
Thanks Derek. Kerryn was just fantastic!
Great to hear that, Hyacinth. Best of luck with your research!
Its a good templates very attractive and important to PhD students and lectuter
Thanks for the feedback, Matobela. Good luck with your research methodology.
Thank you. This is really helpful.
You’re very welcome, Elie. Good luck with your research methodology.
Well explained thanks
This is a very helpful site especially for young researchers at college. It provides sufficient information to guide students and equip them with the necessary foundation to ask any other questions aimed at deepening their understanding.
Thanks for the kind words, Edward. Good luck with your research!
Thank you. I have learned a lot.
Great to hear that, Ngwisa. Good luck with your research methodology!
Thank you for keeping your presentation simples and short and covering key information for research methodology. My key takeaway: Start with defining your research objective the other will depend on the aims of your research question.
My name is Zanele I would like to be assisted with my research , and the topic is shortage of nursing staff globally want are the causes , effects on health, patients and community and also globally
Thanks for making it simple and clear. It greatly helped in understanding research methodology. Regards.
This is well simplified and straight to the point
Thank you Dr
I was given an assignment to research 2 publications and describe their research methodology? I don’t know how to start this task can someone help me?
Sure. You’re welcome to book an initial consultation with one of our Research Coaches to discuss how we can assist – https://gradcoach.com/book/new/ .
Thanks a lot I am relieved of a heavy burden.keep up with the good work
I’m very much grateful Dr Derek. I’m planning to pursue one of the careers that really needs one to be very much eager to know. There’s a lot of research to do and everything, but since I’ve gotten this information I will use it to the best of my potential.
Thank you so much, words are not enough to explain how helpful this session has been for me!
Thanks this has thought me alot.
Very concise and helpful. Thanks a lot
Thank Derek. This is very helpful. Your step by step explanation has made it easier for me to understand different concepts. Now i can get on with my research.
I wish i had come across this sooner. So simple but yet insightful
really nice explanation thank you so much
I’m so grateful finding this site, it’s really helpful…….every term well explained and provide accurate understanding especially to student going into an in-depth research for the very first time, even though my lecturer already explained this topic to the class, I think I got the clear and efficient explanation here, much thanks to the author.
It is very helpful material
I would like to be assisted with my research topic : Literature Review and research methodologies. My topic is : what is the relationship between unemployment and economic growth?
Its really nice and good for us.
THANKS SO MUCH FOR EXPLANATION, ITS VERY CLEAR TO ME WHAT I WILL BE DOING FROM NOW .GREAT READS.
Short but sweet.Thank you
Informative article. Thanks for your detailed information.
I’m currently working on my Ph.D. thesis. Thanks a lot, Derek and Kerryn, Well-organized sequences, facilitate the readers’ following.
great article for someone who does not have any background can even understand
I am a bit confused about research design and methodology. Are they the same? If not, what are the differences and how are they related?
Thanks in advance.
concise and informative.
Thank you very much
How can we site this article is Harvard style?
Very well written piece that afforded better understanding of the concept. Thank you!
Am a new researcher trying to learn how best to write a research proposal. I find your article spot on and want to download the free template but finding difficulties. Can u kindly send it to my email, the free download entitled, “Free Download: Research Proposal Template (with Examples)”.
Thank too much
Thank you very much for your comprehensive explanation about research methodology so I like to thank you again for giving us such great things.
Good very well explained.Thanks for sharing it.
Thank u sir, it is really a good guideline.
so helpful thank you very much.
Thanks for the video it was very explanatory and detailed, easy to comprehend and follow up. please, keep it up the good work
It was very helpful, a well-written document with precise information.
how do i reference this?
MLA Jansen, Derek, and Kerryn Warren. “What (Exactly) Is Research Methodology?” Grad Coach, June 2021, gradcoach.com/what-is-research-methodology/.
APA Jansen, D., & Warren, K. (2021, June). What (Exactly) Is Research Methodology? Grad Coach. https://gradcoach.com/what-is-research-methodology/
Your explanation is easily understood. Thank you
Very help article. Now I can go my methodology chapter in my thesis with ease
I feel guided ,Thank you
This simplification is very helpful. It is simple but very educative, thanks ever so much
The write up is informative and educative. It is an academic intellectual representation that every good researcher can find useful. Thanks
Wow, this is wonderful long live.
Nice initiative
thank you the video was helpful to me.
Thank you very much for your simple and clear explanations I’m really satisfied by the way you did it By now, I think I can realize a very good article by following your fastidious indications May God bless you
Thanks very much, it was very concise and informational for a beginner like me to gain an insight into what i am about to undertake. I really appreciate.
very informative sir, it is amazing to understand the meaning of question hidden behind that, and simple language is used other than legislature to understand easily. stay happy.
This one is really amazing. All content in your youtube channel is a very helpful guide for doing research. Thanks, GradCoach.
research methodologies
Please send me more information concerning dissertation research.
Nice piece of knowledge shared….. #Thump_UP
This is amazing, it has said it all. Thanks to Gradcoach
This is wonderful,very elaborate and clear.I hope to reach out for your assistance in my research very soon.
This is the answer I am searching about…
realy thanks a lot
Thank you very much for this awesome, to the point and inclusive article.
Thank you very much I need validity and reliability explanation I have exams
Thank you for a well explained piece. This will help me going forward.
Very simple and well detailed Many thanks
This is so very simple yet so very effective and comprehensive. An Excellent piece of work.
I wish I saw this earlier on! Great insights for a beginner(researcher) like me. Thanks a mil!
Thank you very much, for such a simplified, clear and practical step by step both for academic students and general research work. Holistic, effective to use and easy to read step by step. One can easily apply the steps in practical terms and produce a quality document/up-to standard
Thanks for simplifying these terms for us, really appreciated.
Thanks for a great work. well understood .
This was very helpful. It was simple but profound and very easy to understand. Thank you so much!
Great and amazing research guidelines. Best site for learning research
hello sir/ma’am, i didn’t find yet that what type of research methodology i am using. because i am writing my report on CSR and collect all my data from websites and articles so which type of methodology i should write in dissertation report. please help me. i am from India.
how does this really work?
perfect content, thanks a lot
As a researcher, I commend you for the detailed and simplified information on the topic in question. I would like to remain in touch for the sharing of research ideas on other topics. Thank you
Impressive. Thank you, Grad Coach 😍
Thank you Grad Coach for this piece of information. I have at least learned about the different types of research methodologies.
Very useful content with easy way
Thank you very much for the presentation. I am an MPH student with the Adventist University of Africa. I have successfully completed my theory and starting on my research this July. My topic is “Factors associated with Dental Caries in (one District) in Botswana. I need help on how to go about this quantitative research
I am so grateful to run across something that was sooo helpful. I have been on my doctorate journey for quite some time. Your breakdown on methodology helped me to refresh my intent. Thank you.
thanks so much for this good lecture. student from university of science and technology, Wudil. Kano Nigeria.
It’s profound easy to understand I appreciate
Thanks a lot for sharing superb information in a detailed but concise manner. It was really helpful and helped a lot in getting into my own research methodology.
Comment * thanks very much
This was sooo helpful for me thank you so much i didn’t even know what i had to write thank you!
You’re most welcome 🙂
Simple and good. Very much helpful. Thank you so much.
This is very good work. I have benefited.
Thank you so much for sharing
This is powerful thank you so much guys
I am nkasa lizwi doing my research proposal on honors with the university of Walter Sisulu Komani I m on part 3 now can you assist me.my topic is: transitional challenges faced by educators in intermediate phase in the Alfred Nzo District.
Appreciate the presentation. Very useful step-by-step guidelines to follow.
I appreciate sir
wow! This is super insightful for me. Thank you!
Indeed this material is very helpful! Kudos writers/authors.
I want to say thank you very much, I got a lot of info and knowledge. Be blessed.
I want present a seminar paper on Optimisation of Deep learning-based models on vulnerability detection in digital transactions.
Need assistance
Dear Sir, I want to be assisted on my research on Sanitation and Water management in emergencies areas.
I am deeply grateful for the knowledge gained. I will be getting in touch shortly as I want to be assisted in my ongoing research.
The information shared is informative, crisp and clear. Kudos Team! And thanks a lot!
hello i want to study
Hello!! Grad coach teams. I am extremely happy in your tutorial or consultation. i am really benefited all material and briefing. Thank you very much for your generous helps. Please keep it up. If you add in your briefing, references for further reading, it will be very nice.
All I have to say is, thank u gyz.
Good, l thanks
thank you, it is very useful
Trackbacks/Pingbacks
- What Is A Literature Review (In A Dissertation Or Thesis) - Grad Coach - […] the literature review is to inform the choice of methodology for your own research. As we’ve discussed on the Grad Coach blog,…
- Free Download: Research Proposal Template (With Examples) - Grad Coach - […] Research design (methodology) […]
- Dissertation vs Thesis: What's the difference? - Grad Coach - […] and thesis writing on a daily basis – everything from how to find a good research topic to which…
Submit a Comment Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
Submit Comment
- Print Friendly

What is Research Methodology? Definition, Types, and Examples

Research methodology 1,2 is a structured and scientific approach used to collect, analyze, and interpret quantitative or qualitative data to answer research questions or test hypotheses. A research methodology is like a plan for carrying out research and helps keep researchers on track by limiting the scope of the research. Several aspects must be considered before selecting an appropriate research methodology, such as research limitations and ethical concerns that may affect your research.
The research methodology section in a scientific paper describes the different methodological choices made, such as the data collection and analysis methods, and why these choices were selected. The reasons should explain why the methods chosen are the most appropriate to answer the research question. A good research methodology also helps ensure the reliability and validity of the research findings. There are three types of research methodology—quantitative, qualitative, and mixed-method, which can be chosen based on the research objectives.
What is research methodology ?
A research methodology describes the techniques and procedures used to identify and analyze information regarding a specific research topic. It is a process by which researchers design their study so that they can achieve their objectives using the selected research instruments. It includes all the important aspects of research, including research design, data collection methods, data analysis methods, and the overall framework within which the research is conducted. While these points can help you understand what is research methodology, you also need to know why it is important to pick the right methodology.
Why is research methodology important?
Having a good research methodology in place has the following advantages: 3
- Helps other researchers who may want to replicate your research; the explanations will be of benefit to them.
- You can easily answer any questions about your research if they arise at a later stage.
- A research methodology provides a framework and guidelines for researchers to clearly define research questions, hypotheses, and objectives.
- It helps researchers identify the most appropriate research design, sampling technique, and data collection and analysis methods.
- A sound research methodology helps researchers ensure that their findings are valid and reliable and free from biases and errors.
- It also helps ensure that ethical guidelines are followed while conducting research.
- A good research methodology helps researchers in planning their research efficiently, by ensuring optimum usage of their time and resources.
Writing the methods section of a research paper? Let Paperpal help you achieve perfection
Types of research methodology.
There are three types of research methodology based on the type of research and the data required. 1
- Quantitative research methodology focuses on measuring and testing numerical data. This approach is good for reaching a large number of people in a short amount of time. This type of research helps in testing the causal relationships between variables, making predictions, and generalizing results to wider populations.
- Qualitative research methodology examines the opinions, behaviors, and experiences of people. It collects and analyzes words and textual data. This research methodology requires fewer participants but is still more time consuming because the time spent per participant is quite large. This method is used in exploratory research where the research problem being investigated is not clearly defined.
- Mixed-method research methodology uses the characteristics of both quantitative and qualitative research methodologies in the same study. This method allows researchers to validate their findings, verify if the results observed using both methods are complementary, and explain any unexpected results obtained from one method by using the other method.
What are the types of sampling designs in research methodology?
Sampling 4 is an important part of a research methodology and involves selecting a representative sample of the population to conduct the study, making statistical inferences about them, and estimating the characteristics of the whole population based on these inferences. There are two types of sampling designs in research methodology—probability and nonprobability.
- Probability sampling
In this type of sampling design, a sample is chosen from a larger population using some form of random selection, that is, every member of the population has an equal chance of being selected. The different types of probability sampling are:
- Systematic —sample members are chosen at regular intervals. It requires selecting a starting point for the sample and sample size determination that can be repeated at regular intervals. This type of sampling method has a predefined range; hence, it is the least time consuming.
- Stratified —researchers divide the population into smaller groups that don’t overlap but represent the entire population. While sampling, these groups can be organized, and then a sample can be drawn from each group separately.
- Cluster —the population is divided into clusters based on demographic parameters like age, sex, location, etc.
- Convenience —selects participants who are most easily accessible to researchers due to geographical proximity, availability at a particular time, etc.
- Purposive —participants are selected at the researcher’s discretion. Researchers consider the purpose of the study and the understanding of the target audience.
- Snowball —already selected participants use their social networks to refer the researcher to other potential participants.
- Quota —while designing the study, the researchers decide how many people with which characteristics to include as participants. The characteristics help in choosing people most likely to provide insights into the subject.
What are data collection methods?
During research, data are collected using various methods depending on the research methodology being followed and the research methods being undertaken. Both qualitative and quantitative research have different data collection methods, as listed below.
Qualitative research 5
- One-on-one interviews: Helps the interviewers understand a respondent’s subjective opinion and experience pertaining to a specific topic or event
- Document study/literature review/record keeping: Researchers’ review of already existing written materials such as archives, annual reports, research articles, guidelines, policy documents, etc.
- Focus groups: Constructive discussions that usually include a small sample of about 6-10 people and a moderator, to understand the participants’ opinion on a given topic.
- Qualitative observation : Researchers collect data using their five senses (sight, smell, touch, taste, and hearing).
Quantitative research 6
- Sampling: The most common type is probability sampling.
- Interviews: Commonly telephonic or done in-person.
- Observations: Structured observations are most commonly used in quantitative research. In this method, researchers make observations about specific behaviors of individuals in a structured setting.
- Document review: Reviewing existing research or documents to collect evidence for supporting the research.
- Surveys and questionnaires. Surveys can be administered both online and offline depending on the requirement and sample size.
Let Paperpal help you write the perfect research methods section. Start now!
What are data analysis methods.
The data collected using the various methods for qualitative and quantitative research need to be analyzed to generate meaningful conclusions. These data analysis methods 7 also differ between quantitative and qualitative research.
Quantitative research involves a deductive method for data analysis where hypotheses are developed at the beginning of the research and precise measurement is required. The methods include statistical analysis applications to analyze numerical data and are grouped into two categories—descriptive and inferential.
Descriptive analysis is used to describe the basic features of different types of data to present it in a way that ensures the patterns become meaningful. The different types of descriptive analysis methods are:
- Measures of frequency (count, percent, frequency)
- Measures of central tendency (mean, median, mode)
- Measures of dispersion or variation (range, variance, standard deviation)
- Measure of position (percentile ranks, quartile ranks)
Inferential analysis is used to make predictions about a larger population based on the analysis of the data collected from a smaller population. This analysis is used to study the relationships between different variables. Some commonly used inferential data analysis methods are:
- Correlation: To understand the relationship between two or more variables.
- Cross-tabulation: Analyze the relationship between multiple variables.
- Regression analysis: Study the impact of independent variables on the dependent variable.
- Frequency tables: To understand the frequency of data.
- Analysis of variance: To test the degree to which two or more variables differ in an experiment.
Qualitative research involves an inductive method for data analysis where hypotheses are developed after data collection. The methods include:
- Content analysis: For analyzing documented information from text and images by determining the presence of certain words or concepts in texts.
- Narrative analysis: For analyzing content obtained from sources such as interviews, field observations, and surveys. The stories and opinions shared by people are used to answer research questions.
- Discourse analysis: For analyzing interactions with people considering the social context, that is, the lifestyle and environment, under which the interaction occurs.
- Grounded theory: Involves hypothesis creation by data collection and analysis to explain why a phenomenon occurred.
- Thematic analysis: To identify important themes or patterns in data and use these to address an issue.
How to choose a research methodology?
Here are some important factors to consider when choosing a research methodology: 8
- Research objectives, aims, and questions —these would help structure the research design.
- Review existing literature to identify any gaps in knowledge.
- Check the statistical requirements —if data-driven or statistical results are needed then quantitative research is the best. If the research questions can be answered based on people’s opinions and perceptions, then qualitative research is most suitable.
- Sample size —sample size can often determine the feasibility of a research methodology. For a large sample, less effort- and time-intensive methods are appropriate.
- Constraints —constraints of time, geography, and resources can help define the appropriate methodology.
Got writer’s block? Kickstart your research paper writing with Paperpal now!
How to write a research methodology .
A research methodology should include the following components: 3,9
- Research design —should be selected based on the research question and the data required. Common research designs include experimental, quasi-experimental, correlational, descriptive, and exploratory.
- Research method —this can be quantitative, qualitative, or mixed-method.
- Reason for selecting a specific methodology —explain why this methodology is the most suitable to answer your research problem.
- Research instruments —explain the research instruments you plan to use, mainly referring to the data collection methods such as interviews, surveys, etc. Here as well, a reason should be mentioned for selecting the particular instrument.
- Sampling —this involves selecting a representative subset of the population being studied.
- Data collection —involves gathering data using several data collection methods, such as surveys, interviews, etc.
- Data analysis —describe the data analysis methods you will use once you’ve collected the data.
- Research limitations —mention any limitations you foresee while conducting your research.
- Validity and reliability —validity helps identify the accuracy and truthfulness of the findings; reliability refers to the consistency and stability of the results over time and across different conditions.
- Ethical considerations —research should be conducted ethically. The considerations include obtaining consent from participants, maintaining confidentiality, and addressing conflicts of interest.
Streamline Your Research Paper Writing Process with Paperpal
The methods section is a critical part of the research papers, allowing researchers to use this to understand your findings and replicate your work when pursuing their own research. However, it is usually also the most difficult section to write. This is where Paperpal can help you overcome the writer’s block and create the first draft in minutes with Paperpal Copilot, its secure generative AI feature suite.
With Paperpal you can get research advice, write and refine your work, rephrase and verify the writing, and ensure submission readiness, all in one place. Here’s how you can use Paperpal to develop the first draft of your methods section.
- Generate an outline: Input some details about your research to instantly generate an outline for your methods section
- Develop the section: Use the outline and suggested sentence templates to expand your ideas and develop the first draft.
- P araph ras e and trim : Get clear, concise academic text with paraphrasing that conveys your work effectively and word reduction to fix redundancies.
- Choose the right words: Enhance text by choosing contextual synonyms based on how the words have been used in previously published work.
- Check and verify text : Make sure the generated text showcases your methods correctly, has all the right citations, and is original and authentic. .
You can repeat this process to develop each section of your research manuscript, including the title, abstract and keywords. Ready to write your research papers faster, better, and without the stress? Sign up for Paperpal and start writing today!
Frequently Asked Questions
Q1. What are the key components of research methodology?
A1. A good research methodology has the following key components:
- Research design
- Data collection procedures
- Data analysis methods
- Ethical considerations
Q2. Why is ethical consideration important in research methodology?
A2. Ethical consideration is important in research methodology to ensure the readers of the reliability and validity of the study. Researchers must clearly mention the ethical norms and standards followed during the conduct of the research and also mention if the research has been cleared by any institutional board. The following 10 points are the important principles related to ethical considerations: 10
- Participants should not be subjected to harm.
- Respect for the dignity of participants should be prioritized.
- Full consent should be obtained from participants before the study.
- Participants’ privacy should be ensured.
- Confidentiality of the research data should be ensured.
- Anonymity of individuals and organizations participating in the research should be maintained.
- The aims and objectives of the research should not be exaggerated.
- Affiliations, sources of funding, and any possible conflicts of interest should be declared.
- Communication in relation to the research should be honest and transparent.
- Misleading information and biased representation of primary data findings should be avoided.
Q3. What is the difference between methodology and method?
A3. Research methodology is different from a research method, although both terms are often confused. Research methods are the tools used to gather data, while the research methodology provides a framework for how research is planned, conducted, and analyzed. The latter guides researchers in making decisions about the most appropriate methods for their research. Research methods refer to the specific techniques, procedures, and tools used by researchers to collect, analyze, and interpret data, for instance surveys, questionnaires, interviews, etc.
Research methodology is, thus, an integral part of a research study. It helps ensure that you stay on track to meet your research objectives and answer your research questions using the most appropriate data collection and analysis tools based on your research design.
Accelerate your research paper writing with Paperpal. Try for free now!
- Research methodologies. Pfeiffer Library website. Accessed August 15, 2023. https://library.tiffin.edu/researchmethodologies/whatareresearchmethodologies
- Types of research methodology. Eduvoice website. Accessed August 16, 2023. https://eduvoice.in/types-research-methodology/
- The basics of research methodology: A key to quality research. Voxco. Accessed August 16, 2023. https://www.voxco.com/blog/what-is-research-methodology/
- Sampling methods: Types with examples. QuestionPro website. Accessed August 16, 2023. https://www.questionpro.com/blog/types-of-sampling-for-social-research/
- What is qualitative research? Methods, types, approaches, examples. Researcher.Life blog. Accessed August 15, 2023. https://researcher.life/blog/article/what-is-qualitative-research-methods-types-examples/
- What is quantitative research? Definition, methods, types, and examples. Researcher.Life blog. Accessed August 15, 2023. https://researcher.life/blog/article/what-is-quantitative-research-types-and-examples/
- Data analysis in research: Types & methods. QuestionPro website. Accessed August 16, 2023. https://www.questionpro.com/blog/data-analysis-in-research/#Data_analysis_in_qualitative_research
- Factors to consider while choosing the right research methodology. PhD Monster website. Accessed August 17, 2023. https://www.phdmonster.com/factors-to-consider-while-choosing-the-right-research-methodology/
- What is research methodology? Research and writing guides. Accessed August 14, 2023. https://paperpile.com/g/what-is-research-methodology/
- Ethical considerations. Business research methodology website. Accessed August 17, 2023. https://research-methodology.net/research-methodology/ethical-considerations/
Paperpal is a comprehensive AI writing toolkit that helps students and researchers achieve 2x the writing in half the time. It leverages 21+ years of STM experience and insights from millions of research articles to provide in-depth academic writing, language editing, and submission readiness support to help you write better, faster.
Get accurate academic translations, rewriting support, grammar checks, vocabulary suggestions, and generative AI assistance that delivers human precision at machine speed. Try for free or upgrade to Paperpal Prime starting at US$19 a month to access premium features, including consistency, plagiarism, and 30+ submission readiness checks to help you succeed.
Experience the future of academic writing – Sign up to Paperpal and start writing for free!
Related Reads:
- Dangling Modifiers and How to Avoid Them in Your Writing
- Webinar: How to Use Generative AI Tools Ethically in Your Academic Writing
- Research Outlines: How to Write An Introduction Section in Minutes with Paperpal Copilot
- How to Paraphrase Research Papers Effectively
Language and Grammar Rules for Academic Writing
Climatic vs. climactic: difference and examples, you may also like, dissertation printing and binding | types & comparison , what is a dissertation preface definition and examples , how to write a research proposal: (with examples..., how to write your research paper in apa..., how to choose a dissertation topic, how to write a phd research proposal, how to write an academic paragraph (step-by-step guide), maintaining academic integrity with paperpal’s generative ai writing..., research funding basics: what should a grant proposal..., how to write an abstract in research papers....
Research Methods
Chapter 2 introduction.
Maybe you have already gained some experience in doing research, for example in your bachelor studies, or as part of your work.
The challenge in conducting academic research at masters level, is that it is multi-faceted.
The types of activities are:
- Finding and reviewing literature on your research topic;
- Designing a research project that will answer your research questions;
- Collecting relevant data from one or more sources;
- Analyzing the data, statistically or otherwise, and
- Writing up and presenting your findings.
Some researchers are strong on some parts but weak on others.
We do not require perfection. But we do require high quality.
Going through all stages of the research project, with the guidance of your supervisor, is a learning process.
The journey is hard at times, but in the end your thesis is considered an academic publication, and we want you to be proud of what you have achieved!
Probably the biggest challenge is, where to begin?
- What will be your topic?
- And once you have selected a topic, what are the questions that you want to answer, and how?
In the first chapter of the book, you will find several views on the nature and scope of business research.
Since a study in business administration derives its relevance from its application to real-life situations, an MBA typically falls in the grey area between applied research and basic research.
The focus of applied research is on finding solutions to problems, and on improving (y)our understanding of existing theories of management.
Applied research that makes use of existing theories, often leads to amendments or refinements of these theories. That is, the applied research feeds back to basic research.
In the early stages of your research, you will feel like you are running around in circles.
You start with an idea for a research topic. Then, after reading literature on the topic, you will revise or refine your idea. And start reading again with a clearer focus ...
A thesis research/project typically consists of two main stages.
The first stage is the research proposal .
Once the research proposal has been approved, you can start with the data collection, analysis and write-up (including conclusions and recommendations).
Stage 1, the research proposal consists of he first three chapters of the commonly used five-chapter structure :
- Chapter 1: Introduction
- An introduction to the topic.
- The research questions that you want to answer (and/or hypotheses that you want to test).
- A note on why the research is of academic and/or professional relevance.
- Chapter 2: Literature
- A review of relevant literature on the topic.
- Chapter 3: Methodology
The methodology is at the core of your research. Here, you define how you are going to do the research. What data will be collected, and how?
Your data should allow you to answer your research questions. In the research proposal, you will also provide answers to the questions when and how much . Is it feasible to conduct the research within the given time-frame (say, 3-6 months for a typical master thesis)? And do you have the resources to collect and analyze the data?
In stage 2 you collect and analyze the data, and write the conclusions.
- Chapter 4: Data Analysis and Findings
- Chapter 5: Summary, Conclusions and Recommendations
This video gives a nice overview of the elements of writing a thesis.
Academia.edu no longer supports Internet Explorer.
To browse Academia.edu and the wider internet faster and more securely, please take a few seconds to upgrade your browser .
Enter the email address you signed up with and we'll email you a reset link.
- We're Hiring!
- Help Center

CHAPTER THREE RESEARCH METHODOLOGY Introduction

Related Papers
Golden Ratio Of Social Science And Education
Michael Mncedisi Willie
Mercy Ndoro
Management Studies ISSN 2328-2185
Nidhima gupta
Nazibur Rahman
International Journal of Business and Technopreneurship (IJBT)
Nor Siah Jaharuddin
The study underscores the paramount importance of meticulous population selection and sampling strategy in research design. Providing researchers with a comprehensive overview of population considerations and sampling methods, it offers a valuable resource for enhancing the robustness and applicability of research outcomes across diverse disciplines. Researchers discuss the unit of analysis, unit of observation, population of interest, target population, sampling framework, and sampling methods in light of employee work engagement in Malaysia. Simple random sampling, stratified random sampling, systematic random sampling, cluster sampling (single-stage, double-stage, and multi-stage), phase sampling (two-phase and multiphase), convenience sampling, purposive sampling, quota sampling, snowball sampling, and volunteer sampling have been discussed for selecting the appropriate sampling method for the research titled Revisiting of JD-R Theory and the effect of leadership style and meani...
Muhammad Saadi
introduction to business statistics
Erana Terefa
Population: aggregation of the elements from which a sample is actually selected. It is the entire group of individuals or objects under consideration. Sample: it is a subgroup or part of the population selected by some method in order estimate population characteristics. Elementary unit (unit of analysis): an element or group of elements on which information is required or it is the object that we observe or measure. Thus, persons, vehicles, households, farms are examples of elementary units. Sampling units: for the purpose of sample selection, the population is divided in to a finite number of distinct, non-overlapping and identifiable units called sampling units. Sample Frame: is a list of elements covering the survey population, and serves as a base for sample selection. Data: These are measurements or observations (values) recorded for each element. Variable: is a characteristic or attribute that can assume different values. Population parameters: These are facts about population/descriptions of population. Statistic: it is characteristic or a fact about a sample. THE NEED FOR SAMPLING: The following points summarize the benefits of studying samples. Sampling can save time and money. A sample study is usually less expensive than a census study and produces results at a relatively faster speed. There could be resource (time, finance, manpower, etc.) limitations which would make it difficult to study the whole population. Sampling may enable more accurate measurements for a sample study is generally conducted by trained and experienced investigators Business statistics Chapter-1-1. SAMPLING AND SAMPLING DISTRIBUTIONS
Alliana Ulila
Loading Preview
Sorry, preview is currently unavailable. You can download the paper by clicking the button above.
RELATED PAPERS
Obaidullah Khan
KABIR TAHIR HAMID
Godwill Medukenya
Organisational Culture and Employee Engagement in the Banking Industry in Benin City
Samuel J . Osifo (Ph.D)
Mohamed Adam, Hassan
Quantitative Social Research Methods
kultar singh
Journal of Resources Development and Management: 30-41
NWOKWU, P . M., PhD
Ayhan Nadiri
Complexity Perspectives in Innovation and Social Change (D. Lane et al. (eds.)
Dwight Read
De Wet Schutte
Alexis Hernandez
DGeanene White
International Journal Of Medical Science And Clinical Research Studies
Michael Willie
Dustin Johanna Satterfield
ACM SIGSOFT Software Engineering Notes
Aris Munandar
estime.ird.fr
Rigas Arvanitis
Abebe T Kassaye
Adnan Hussain
Associate Professor Dr. Sanya - Kenaphoom
Dr. A. Michael Masilamani
KIU Journal of Social Sciences , Oyetola Oniwide
ezekiel nnadi
Bhupender Pandey
- We're Hiring!
- Help Center
- Find new research papers in:
- Health Sciences
- Earth Sciences
- Cognitive Science
- Mathematics
- Computer Science
- Academia ©2024

Style and Grammar Guidelines
APA Style provides a foundation for effective scholarly communication because it helps writers present their ideas in a clear, concise, and inclusive manner. When style works best, ideas flow logically, sources are credited appropriately, and papers are organized predictably. People are described using language that affirms their worth and dignity. Authors plan for ethical compliance and report critical details of their research protocol to allow readers to evaluate findings and other researchers to potentially replicate the studies. Tables and figures present information in an engaging, readable manner.
The style and grammar guidelines pages present information about APA Style as described in the Publication Manual of the American Psychological Association, Seventh Edition and the Concise Guide to APA Style, Seventh Edition . Any updates to APA Style are noted on the applicable topic pages. If you are still using the sixth edition, helpful resources are available in the sixth edition archive .
Looking for more style?

- Accessibility of APA Style
- Line Spacing
- Order of Pages
- Page Header
- Paragraph Alignment and Indentation
- Sample Papers
- Title Page Setup
- Appropriate Level of Citation
- Basic Principles of Citation
- Classroom or Intranet Sources
- Paraphrasing
- Personal Communications
- Quotations From Research Participants
- Secondary Sources
- Abbreviations
- Capitalization
- Italics and Quotation Marks
- Punctuation
- Spelling and Hyphenation
- General Principles for Reducing Bias
- Historical Context
- Intersectionality
- Participation in Research
- Racial and Ethnic Identity
- Sexual Orientation
- Socioeconomic Status
- Accessible Use of Color in Figures
- Figure Setup
- Sample Figures
- Sample Tables
- Table Setup
- Archival Documents and Collections
- Basic Principles of Reference List Entries
- Database Information in References
- DOIs and URLs
- Elements of Reference List Entries
- Missing Reference Information
- Reference Examples
- References in a Meta-Analysis
- Reference Lists Versus Bibliographies
- Works Included in a Reference List
- Active and Passive Voice
- Anthropomorphism
- First-Person Pronouns
- Logical Comparisons
- Plural Nouns
- Possessive Adjectives
- Possessive Nouns
- Singular “They”
- Adapting a Dissertation or Thesis Into a Journal Article
- Correction Notices
- Cover Letters
- Journal Article Reporting Standards (JARS)
- Open Science
- Response to Reviewers
- Open access
- Published: 26 August 2024
Inter-laboratory comparison of eleven quantitative or digital PCR assays for detection of proviral bovine leukemia virus in blood samples
- Aneta Pluta 1 , 13 ,
- Juan Pablo Jaworski 2 ,
- Casey Droscha 3 ,
- Sophie VanderWeele 3 ,
- Tasia M. Taxis 4 ,
- Stephen Valas 5 ,
- Dragan Brnić 6 ,
- Andreja Jungić 6 ,
- María José Ruano 7 ,
- Azucena Sánchez 7 ,
- Kenji Murakami 8 ,
- Kurumi Nakamura 8 ,
- Rodrigo Puentes 9 ,
- MLaureana De Brun 9 ,
- Vanesa Ruiz 2 ,
- Marla Eliana Ladera Gómez 10 ,
- Pamela Lendez 10 ,
- Guillermina Dolcini 10 ,
- Marcelo Fernandes Camargos 11 ,
- Antônio Fonseca 11 ,
- Subarna Barua 12 ,
- Chengming Wang 12 ,
- Aleksandra Giza 13 &
- Jacek Kuźmak 1
BMC Veterinary Research volume 20 , Article number: 381 ( 2024 ) Cite this article
154 Accesses
1 Altmetric
Metrics details
Bovine leukemia virus (BLV) is the etiological agent of enzootic bovine leukosis and causes a persistent infection that can leave cattle with no symptoms. Many countries have been able to successfully eradicate BLV through improved detection and management methods. However, with the increasing novel molecular detection methods there have been few efforts to standardize these results at global scale. This study aimed to determine the interlaboratory accuracy and agreement of 11 molecular tests in detecting BLV. Each qPCR/ddPCR method varied by target gene, primer design, DNA input and chemistries. DNA samples were extracted from blood of BLV-seropositive cattle and lyophilized to grant a better preservation during shipping to all participants around the globe. Twenty nine out of 44 samples were correctly identified by the 11 labs and all methods exhibited a diagnostic sensitivity between 74 and 100%. Agreement amongst different assays was linked to BLV copy numbers present in samples and the characteristics of each assay (i.e., BLV target sequence). Finally, the mean correlation value for all assays was within the range of strong correlation. This study highlights the importance of continuous need for standardization and harmonization amongst assays and the different participants. The results underscore the need of an international calibrator to estimate the efficiency (standard curve) of the different assays and improve quantitation accuracy. Additionally, this will inform future participants about the variability associated with emerging chemistries, methods, and technologies used to study BLV. Altogether, by improving tests performance worldwide it will positively aid in the eradication efforts.
Peer Review reports
Introduction
Bovine leukemia virus (BLV) is a deltaretrovirus from the Orthoretrovirinae subfamily of the Retroviridae family. An essential step in the BLV replication cycle is the integration of DNA copy of its RNA genome into the DNA of a host cell [ 1 ]. Once integrated, the proviral DNA is replicated along with the host’s DNA during cellular divisions, as for any cellular gene. The BLV is the etiologic agent of enzootic bovine leukosis (EBL). BLV causes a persistent infection in cattle, and in most cases this infection is asymptomatic [ 2 ]. In one-third of infected animals the infection progresses to a state of persistent lymphocytosis, and in 1 to 10% of infected cattle it develops into lymphosarcoma [ 2 ]. BLV induces high economic losses due to trade restrictions, replacement cost, reduced milk production, immunosuppression, and increased susceptibility to pneumonia, diarrhea, mastitis, and so on [ 3 , 4 , 5 , 6 ]. BLV is globally distributed with a high prevalence, except for Western Europe and Oceania, where the virus has been successfully eradicated through detection and elimination of BLV-infected animals [ 7 , 8 ]. The agar gel immunodiffusion and ELISA for the detection of BLV-specific antibodies in sera and milk are the World Organization for Animal Health (WOAH, founded as OIE) prescribed tests for serological diagnosis but ELISA, due to its high sensitivity and ability to test many samples at a very low cost, is highly recommended [ 9 ]. Despite the advantages of serologic testing, there are some scenarios in which direct detection of the BLV genomic fragment was important to improve BLV detection. The most frequent cases is the screening of calves with maternal antibodies, acute infection, animals without persistent antibody response and animal subproducts (i.e., semen). In this regard, nucleic acid amplification tests such as real-time quantitative PCR (qPCR) allows for a rapid and highly sensitive detection of BLV proviral DNA (BLV DNA) that can be used to test infected and asymptomatic animals, before the elicitation of anti-BLV specific antibodies and when proviral load (PVL) are still low [ 10 ]. Furthermore, qPCR assays can serve as confirmatory tests for the clarification of inconclusive and discordant serological test results usually associated with these cases [ 11 ]. For these reasons, the inclusion of qPCR in combination with other screening tests might increase control programs efficiency. Additionally, qPCR allows the estimation of BLV PVL which is important for studying the dynamics of BLV infection (i.e., basic research). Further, considering that BLV PVL correlates with the risk of BLV transmission, this feature of qPCR can be exploited for developing rational segregation programs [ 12 , 13 ]. The results of Kobayashi et al. suggest that high PVL is also a significant risk factor for progression to EBL and should therefore be used as a parameter to identify cattle for culling from the herd well before EBL progression [ 14 ]. Several qPCRs have been developed globally for the quantitation of BLV DNA. Although most assays have been properly validated by each developer, a proper standardization and harmonization of such tests is currently lacking. Considering that standardization and harmonization of qPCR methods and results are essential for comparisons of data from BLV laboratories around the world, this could directly impact international surveillance programs and collaborative research. We built a global collaborative network of BLV reference laboratories to evaluate the interlaboratory variability of different qPCRs and sponsored a harmonization of assays to hopefully impact international surveillance programs and research going forward.
In 2018 we conducted the first global trial of this kind to assess the interlaboratory variability of six qPCRs for the detection of BLV DNA [ 15 ]. Since this complex process is a continuous rather than a one-time effort, we now started a second study of this type. In this follow up study, we built a more comprehensive sample panel, accounting for a broader geographical diversification. Additionally, we increased the number of participants to ten collaborating laboratories plus one WOAH reference lab and tested novel methodologies including digital PCR (ddPCR) and FRET-qPCR. Finally, we established the next steps towards the international standardization of molecular assays for the detection of BLV DNA.
Materials and methods
Participants.
The eleven laboratories that took part in the study were:(i) the Auburn University College of Veterinary Medicine (Auburn, Alabama, United States): (ii) AntelBio, a division of CentralStar Cooperative (Michigan, United States); (iii) Laboratórios Federais de Defesa Agropecuária de Minas Gerais (LFDA-MG, Pedro Leopoldo, Brasil); (iv) Centro de Investigación Veterinaria de Tandil (CIVETAN, Buenos Aires, Argentina); (v) the Faculty of Agriculture Iwate University (Iwate, Japan); (vi) Universidad de la República de Uruguay (UdelaR, Montevideo, Uruguay); (vii) the Croatian Veterinary Institute (Zagreb, Croatia); (viii) Instituto Nacional de Tecnología Agropecuaria (INTA, Buenos Aires, Argentina); (ix) Laboratorio Central de Veterinaria (LCV, Madrid, Spain); (x) the National Veterinary Research Institute (NVRI, Puławy, Poland) and (xi) the French Agency for Food, Environmental and Occupational Health and Safety (Anses, Niort, France). All European laboratories participating in this study are acting as national reference laboratories for EBL, NVRI acts as WOAH reference laboratory for EBL, while the remaining laboratories are nationally renowned entities for BLV diagnostics. The eleven participating methods are referred to below as qPCR1 – qPCR5, ddPCR6, qPCR7 – qPCR11, respectively.
Sample collection and DNA extraction
A total of 42 DNA samples obtained from blood of naturally BLV-infected dairy cattle from Poland, Moldova, Pakistan, Ukraine, Canada and United States were used for this study. Thirty-six of them were archival DNA samples obtained between 2012–2018 as described in our previous studies on samples from Poland ( n = 21) [ 16 , 17 ], Moldova ( n = 4) [ 18 ], Pakistan ( n = 5) [ 19 ] and Ukraine ( n = 6) [ 15 , 20 ]. Between 2020–2021 6 peripheral blood and serum samples from naturally BLV-infected cattle were obtained from three dairy farms of Alberta, Canada and two dairy farms of Michigan, US. Serological testing and sample processing were conducted by the laboratories from which the samples originated. The genomic DNA from Canadian and US samples was extracted from whole blood using a Quick DNA Miniprep Plus kit (Zymo Research) and a DNeasy Blood & Tissue Kit (Qiagen), respectively in University of Calgary and Michigan State University and sent to the NVRI in the form of DNA solutions. Additionally, one plasmid DNA sample (pBLV344) was kindly supplied by Luc Willems (University of Liège, Belgium) and DNA extracted from FLK-BLV cells were included as positive controls. Finally, DNA extracted from PBL of a serologically negative cattle was included as negative control. At the NVRI, the DNA concentration in all samples was estimated by spectrophotometry using a NanoPhotometer (Implen). Each sample was divided into eleven identical aliquots containing between 800 and 4,000 ng of lyophilised genomic DNA. Eleven identical sets of these samples were lyophilized (Alpha 1–4 LSC basic, Martin Christ Gefriertrocknungsanlagen GmbH) and distributed to participating laboratories. At the NVRI, all samples were coded (identification [ 21 ] run numbers 1 to 44) to perform a blinded testing. The samples, together with instructions for their preparation (Additional file 1), were shipped by air at room temperature (RT).
Examination of DNA quality/stability
Since different extraction methods and lyophilization process were employed for the preparation of the DNA samples, it was necessary to test the quality of the DNA at the NVRI laboratory. For that purpose, one complete set of samples ( n = 44) was tested by Fragment Analyzer (Agilent Technologies), before and after freeze-drying, to assess DNA quality by calculating a Genomic Quality Number (GQN) for every sample. Low GQN value (< 2.5) represents sheared or degraded DNA. A high GQN (> 9) represents undegraded DNA. In addition, quality of DNA was assessed by determination of copy number of the histone H3 family 3A ( H3F3A ) housekeeping gene using quantitative real-time PCR (qPCR) [ 22 ]. The qPCR results were expressed as the number of H3F3A gene copies per 300 ng of DNA in each sample. Grubbs´ test was performed to determine outliers. To test the stability of DNA, samples were stored for 20 days at RT (10 days) and at + 4 °C (10 days) and were retested by Fragment Analyzer and qPCR 21 days later. A Mann–Whitney U-test was used to compare the median values between fresh and stored samples (time 0 and time 1), respectively.
Description of BLV qPCR protocols used by participating laboratories
All participating laboratories performed their qPCR or ddPCR using a variety of different equipment, reagents, and reaction conditions, which had been set up, validated, and evaluated previously and are currently used as working protocols. The specific features of each of these protocols are described below and summarized in Table 1 .
All laboratories applied standard procedures for avoiding false-positive results indicative of DNA contamination, such as the use of separate rooms for preparing reaction mixtures, adding the samples, and performing the amplification reaction. One of the ten BLV qPCRs used LTR region and the remaining nine qPCRs used the pol gene as the target sequence for amplification, while the ddPCR amplified the env gene.
Method qPCR1
The BLV qPCR amplifying a 187-bp pol gene was performed according to a previously published methods [ 23 , 24 ]. A real-time fluorescence resonance energy transfer (FRET) PCR was carried out in a 20-μl PCR mixture containing 10 μl handmade reaction master mix and 10 μl genomic DNA. The PCR buffer was 4.5 mM MgCl2, 50 mM KCl, 20 mM Tris–HCl, pH 8.4, supplemented with 0.05% each Tween20 and Non-idet P-40, and 0.03% acetylated BSA (Roche Applied Science). For each 20 μl total reaction volume, the nucleotides were used at 0.2 mM each and 1.5 U Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA) was used. Primers were used at 1 μM, LCRed640 probe was used at 0.2 μM, and 6-FAM probe was used at 0.1 μM. Amplification was performed in the Roche Light Cycler 480 II (Roche Molecular Biochemicals) using 10 min denaturation step at 95 °C, followed by 18 high-stringency step-down thermal cycles and 30 low-stringency fluorescence acquisition cycles.
A plasmid containing the BLV-PCR amplicon region was diluted ten-fold from 1 × 10 5 copies to 10 copies per 10 µl and was used as a standard to measure the BLV copy numbers.
Method qPCR2
A BLV proviral load qPCR assay developed by AntelBio, a division of CentralStar Cooperative Inc. on Applied Biosystems 7500 Real-Time PCR system [ 25 , 33 ]. This multiplex assay amplifies the BLV pol gene along with the bovine β-actin gene and an internal amplification control, “Spike”. A quantitative TaqMan PCR was carried out in a 25-μl PCR mixture containing 12.5 µl of 2X InhibiTaq Multiplex HotStart qPCR MasterMix (Empirical Bioscience), 16 nM each BLV primer, 16 nM each β-actin primer, 8 nM each spike primer, 8 nM BLV FAM-probe, 8 nM β-actin Cy5-probe, 4 nM spike JOE-probe, 1 µl of an internal spike-in control (10,000 copies per µl), 7.25 µl of nuclease-free water and 4 µl of DNA sample for each qPCR reaction. The thermal PCR protocol was as follows: 95 °C for 10 min, 40 × (95 °C for 15 s, 60 °C for 1 min). Copy numbers of both the BLV pol gene and bovine β-Actin were derived using a plasmid containing target sequences, quantified by ddPCR, diluted 1 × 10 6 copies per µl to 10 copies per µl in tenfold dilutions. DNA concentrations of each sample were measured using a Qubit 4 Fluorometer and used in combination with the qPCR copy numbers to calculate BLV copies per 100 ng.
Method qPCR3
The qPCR assays for the BLV LTR gene were performed according to a previously published methods [ 26 ]. Genomic DNA was amplified by TaqMan PCR with 10 μl of GoTaq Probe qPCR Master Mix × 2 (Promega), 0.6 pmol/μl each primer, 0.3 pmol/µl double-quenched probe and 100 ng genomic DNA. Amplification was performed in the CFX96 cycler (BioRad) according to the protocol: 5 min denaturation at 95°C followed by 45 cycles (60 s at 94°C and 60 s at 60°C). The efficiency of each reaction was calculated from the serial dilution of DNA extracted from BLV persistently infected fetal lamb kidney (FLK) cells, starting at a concentration of 100 ng/µl [ 21 ]. The detection limit was tested using a plasmid containing the target of the qPCRs, starting at 10 3 ng/µl.
Method qPCR4
The quantitative real-time PCR was done with the primers for the BLV pol gene as previously described [ 34 ]. The qPCR reaction mix contained 1 × PCR Master Mix with SYBR Green (FastStart Universal SYBR Green Master Rox, Roche), 0.3 μM each primer and 30 ng of extracted genomic DNA. Amplification was performed in QuantStudio 5 Real-Time PCR System (Applied Biosystems) under the following conditions: 2 min at 50 °C, 10 min at 95 °C, 40 cycles of 15 s at 95 °C and 60 s at 60 °C. A standard curve of six tenfold serial dilutions of pBLV, containing 1 × 10 6 to 10 BLV copies, was built and run 3 times for validation of the method. The number of provirus copies per reaction (100 ng) was calculated.
Method qPCR5
BLV PVLs were determined by using qPCR kit, RC202 (Takara Bio, Shiga, Japan) [ 28 , 35 ]. This qPCR assay amplifies the BLV pol gene along with the bovine RPPH1 gene as an internal control. Briefly, 100 ng genomic DNA was amplified by TaqMan PCR with four primers for pol gene and RPPH1 gene according to the manufacturer’s instructions: 30 s denaturation at 95 °C followed by 45 cycles (5 s at 95 °C and 30 s at 60 °C). The qPCR was performed on a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific K.K., Tokyo, Japan). Standard curve was generated by creating tenfold serial dilutions of the standard plasmid included in the kit. The standards for calibration ranged from 1 to 10 6 copies/reaction and were run in duplicate. The number of provirus copies per 100 ng was calculated.

Method ddPCR6
The digital droplet PCR (ddPCR) assay for the env gene of the BLV was performed using the protocol previously described by [ 28 , 29 ]. An absolute quantification by TaqMan ddPCR was performed in a typical 20-μl assay, 1 μl of DNA sample was mixed with 1 μl of each primer (10 μM), 0.5 μl of probe (10 μM), and 2 × Supermix emulsified with oil (Bio-Rad). The droplets were transferred to a 96-well plate (Eppendorf). The PCR assay was performed in a thermocycler (C1000 touch cycler; Bio-Rad) with the following parameters: initial denaturation of 10 min at 95 °C, then 40 cycles of 30 s at 94 °C, and 1 min at 58 °C, with final deactivation of the enzyme for 10 min at 98 °C. The presence of fluorescent droplets determined the number of resulting positive events that were analyzed in the software (QuantaSoft v.1.7.4; Bio-Rad), using dot charts. The number of provirus copies per 100 ng were calculated. Each sample was run in duplicate, and results were averaged.
Method qPCR7
This qPCR method for the BLV pol gene is a modified option of widely available quantitative TaqMan qPCR described by Rola-Łuszczak et al. [ 11 ], using the same primers and standards. A quantitative TaqMan PCR was performed in a 20 μl PCR mix containing 10 μl of 2 × ORA qPCR Probe ROX L Mix (highQu, Kraichtal, Germany), 2 μl primer/probe mix (final concentration 400 nM of each of the primers, 200 nM of BLV probe), and 3 μl extracted genomic DNA. Amplification was performed in the Rotor-Gene Q system (Qiagen) with an initial denaturation step and polymerase activation at 95 °C for 3 min, followed by 45 cycles of 95 °C for 5 s and 60 °C for 30 s. As a standard, plasmid pBLV1 (NVRI, Pulawy, PL) containing a BLV pol fragment was used. Tenfold dilutions of plasmid DNA were made from 1 × 10 10 copies to 1 × 10 1 copies per reaction and used to generate the standard curve and estimate BLV copy number per 100 ng.
Method qPCR8
Proviral load quantification was assessed by SYBR Green real-time quantitative PCR (qPCR) using the pol gene as the target sequence [ 36 ]. Briefly, 12-μl PCR mixture contained Fast Start Universal SYBR Green Master Mix (Roche), 800 nM each BLV pol primers and 1 µl DNA as template. The reactions were incubated at 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s, 55 °C for 15 s and 60 °C for 1 min. All samples were tested in duplicate on a StepOne Plus machine (Applied Biosystems). A positive and negative control, as well as a no-template control, were included in each plate. After the reaction was completed, the specificity of the amplicons was checked by analyzing the individual dissociation curves. As a standard, plasmid pBLV1 (NVRI, Pulawy, PL) containing a BLV pol fragment was used. Tenfold dilutions of plasmid DNA were made from 1 × 10 6 to 10 copies per µl and used to generate the standard curve and estimate BLV copy number per 100 ng.
Method qPCR9
This qPCR method is a modified option of widely available quantitative TaqMan qPCR described by Rola-Łuszczak et al. [ 11 ], using the same primers and standards. The detection of BLV genome was combined with an endogenous control system (Toussaint 2007) in a duplex assay. Briefly, 20-µl qPCR reaction contained AhPath ID™ One-Step RT-PCR Reagents with ROX (Applied Biosystems, CA, USA) – 10 µl of 2 × RT-PCR buffer and 0.8 µl of 25 × RT-PCR enzyme mix, 400 nM each primer for pol gene, 100 nM BLV specific probe, 40 nM each β-actin primer, 40 nM β-actin specific probe and 2 µl DNA sample. All samples were tested in ABI7500 Real-Time PCR System (Applied Biosystems) according to the following protocol: 10 min at 48 °C (reverse transcription), 10 min at 95 °C (inactivation reverse transcriptase / activation Taq polymerase) followed by 45 cycles (15 s at 95 °C and 60 s at 60 °C). As a standard, plasmid pBLV1 (NVRI, Pulawy, PL) containing a BLV pol fragment was used. Tenfold dilutions of plasmid DNA were made from 1 × 10 4 copies to 0.1 copies per μl and used to generate the standard curve and estimate BLV copy number per 100 ng.
Method qPCR10
The BLV qPCR was performed as published previously [ 11 ]. A quantitative TaqMan PCR was carried out in a 25-μl PCR mixture containing 12.5 μl of 2 × QuantiTect Multiplex PCR NoROX master mix (Qiagen), 0.4 μM each primer, 0.2 μM specific BLV probe, and 500 ng of extracted genomic DNA. Amplification was performed in the Rotor-Gene Q system (Qiagen) using an initial denaturation step and polymerase activation at 95 °C for 15 min, followed by 50 cycles of 94 °C for 60 s and 60 °C for 60 s. All samples were amplified in duplicate. As a standard, the pBLV1 plasmid (NVRI, Pulawy, PL), containing a 120-bp BLV pol fragment, was used. Tenfold dilutions of this standard were made from 1 × 10 6 copies per μl to 100 copies per μl and were used to estimate the BLV copy numbers per 100 ng.
Method qPCR11
This qPCR method for the BLV pol gene is a modified option of widely available quantitative TaqMan qPCR described by Rola-Łuszczak et al. [ 11 ], using the same primers and standards. The reaction mixture contained 400 nM of each primer, 200 nM of probe, 10 µl of 2 × SsoFast probes supermix (Bio-Rad), 5 µl of DNA sample and H 2 O up to 20 µl of the final volume. PCR assays were carried out on a CFX96 thermocycler (Bio-Rad) under the following amplification profile: 98 °C for 3 min, followed by 45 cycles of 95 °C for 5 s and 60 °C for 30 s. As a standard, plasmid pBLV1 (NVRI, Pulawy, PL) containing a BLV pol fragment was used. Tenfold dilutions of plasmid DNA were used to generate the standard curve and estimate BLV copy number per 100 ng.
Analysis of BLV pol, env and LTR sequences targeted by particular qPCR/ddPCR assays
In order to assess full-length pol , env and LTR sequence variability among BLV genotypes, all BLV sequences ( n = 2191) available on 30 September 2023 in GenBank ( https://www.ncbi.nlm.nih.gov/GenBank/ ) repository were retrieved. From the collected sequences, 100 pol , env and LTR sequences, which were characterized by the highest level of sequence variability and divergence, were selected for the further analysis. A pol -based, env -based and LTR-based maximum likelihood (ML) phylogenetic trees (see Additional file 6) was constructed to assign genotypes to the unassigned BLV genomes [ 37 , 38 , 39 ]. For all genes and LTR region the Tamura-Nei model and Bootstrap replications (1,000) were applied. In this analysis, pol sequences were assigned to 7 BLV genotypes (G1, G2, G3, G4, G6, G9, and G10), while env and LTR sequences were assigned to 10 BLV genotypes (G1, G2, G3, G4, G5, G6, G7, G8, G9, and G10). Phylogeny of the same isolates assigned to particular genotypes by ML method was confirmed by Mr. Bayes analysis [ 40 , 41 , 42 ] (data not shown). From this analysis, a total of 100 full-length pol, env and LTR sequences were used for multiple-sequence alignment (MSA) using ClustalW algorithm, implemented in MEGA X. For all sequences, nucleotide diversity (π), defined as the average number of nucleotide differences per site between two DNA sequences in all possible pairs in the sample population, was estimated using MEGA X. To measure the relative variation in different positions of aligned genes and LTR region the Shannon’s entropy (a quantitative measure of diversity in the alignment, where H = 0 indicates complete conservation) was estimated using BioEdit v. 7.2.5 software 64. The statistical analyses were performed using DATAtab e.U. Graz, Austria and GraphPad Software by Dotmatics, Boston.
Examination of the quality and stability of DNA samples
To test the quality of DNA samples, the H3F3A copy number of each individual sample was assessed by qPCR at the NVRI. Copy numbers were normalized to DNA mass input and results were expressed as copy numbers per 300 ng of total DNA. The respective values were tested by Grubbs' test. The results for 43 DNA samples (sample ID: 42 with BLV genome plasmid was excluded) followed a normal distribution (Shapiro–Wilk 0.97; P = 0.286), with a mean value of 35,626 copies (95% confidence interval [ 43 ] 33,843 to 37,408 copies), a minimum value of 19,848 copies and a maximum value of 46,951 copies (see Additional file 2). Despite a low value for sample ID: 40 no significant outlier was detected in the dataset ( P > 0.05). Therefore, it can be assumed that the DNA quality was acceptable for all samples present in the panel. Next, DNA stability was assessed by retesting the H3F3A copy numbers in each sample ( n = 43) after a combined storage consisting in 10 days at RT and 10 days at + 4°C. A Mann–Whitney U-test was used to compare the median values between fresh and stored samples (time 0 and time 1, respectively), and no significant difference was observed at the 5% level ( P = 0.187) (Fig. 1 A).
Assessment of the stability of DNA samples. A Shown are copy numbers of the H3F3A housekeeping gene in 43 DNA samples that were stored in 10 days at RT and 10 days at + 4°C and tested twice with a 21-day interval. A Mann–Whitney U-test was used to compare the median values between two groups ( P = 0.187); B Shown are GQN values ( n = 43) tested twice with a 21-day interval: `before freeze-drying` and `after freeze-drying`. A Mann–Whitney U-test results between two groups ( P = 0.236)
In addition, the quality of DNA samples after lyophilization was analyzed. DNA from individual samples ( n = 43) was assessed with the genomic DNA quality number on the Fragment Analyzer system. The GQN from all lyophilized samples ranged from 4.0 to 9.7—that represented undegraded DNA. There was no significant difference in GQN values between `before freeze-drying` and `after freeze-drying` groups with respect to the corresponding DNA samples ( P = 0.236) (Fig. 1 B). Altogether, these results suggested that sample storage, lyophilization and shipping has a minimal impact in DNA stability and further testing during the interlaboratory trial.
Detection of BLV proviral DNA by different qPCR assays
A total of 44 DNA samples, including two positive (ID: 42 and 43) and one negative (ID: 32) controls, were blinded and independently tested by eleven laboratories using their own qPCR methods (Table 2 ). All laboratories measured the concentration of DNA in samples (Additional file 3). BLV provirus copy number was normalized to DNA concentration and expressed per 100 ng of genomic DNA for each test.
Except for the positive (pBLV344 and FLK cell line) and the negative controls, all samples had previously shown detectable levels of BLV-specific antibodies (BLV-Abs) by enzyme-linked immunosorbent assays (ELISA). During the current interlaboratory study, both the positive and negative controls were assessed adequately by all eleven PCR tests. Of all 43 positive samples, 43, 35, 37, 36, 40, 32, 40, 42, 42, 42 and 41 samples were detected as positive by the qPCR1, qPCR2, qPCR3, qPCR4, qPCR5, ddPCR6, qPCR7, qPCR8, qPCR9, qPCR10 and qPCR11 methods, respectively. Based on these observations, the most sensitive method was the qPCR1, and the method with the lowest sensitivity was the ddPCR6. Twenty-nine out of 44 samples were identified correctly by all qPCRs. The remaining 15 samples gave discordant results. Comparison of qualitative results (positive versus negative) from all eleven methods revealed 87.33% overall agreement and a kappa value of 0.396 (Cohen's kappa method adapted by Fleiss) [ 44 , 45 ]. The levels of agreement among the results from the eleven methods are represented in Table 3 . The maximum agreement was seen between two methods (qPCR9 and qPCR10 [100% agreement and a Cohen's kappa value of 1.000]) that used similar protocols and targeted the same region of BLV pol .
Analysis of BLV pol, env and LTR sequences targeted by particular PCR assays
Due to differences in performance observed among the pol -based qPCR assays (the qPCR1, qPCR2, qPCR4, qPCR5 and qPCR7- qPCR11 methods), and considering that the env -based ddPCR6 and LTR-based qPCR3 assay showed the lowest sensitivity and the poorest agreement with the other assays, the degree of sequence variability between the pol , env and LTR genes was addressed. From the MSAs for pol , env and LTR, the nucleotide diversity (π) was calculated. The π value for pol gene was lower than that for LTR and env gene (π pol , 0.023 [standard deviation {SD}, 0.018]; π LTR , 0.024 [SD, 0.011]; π env , 0.037 [SD, 0.013]). From this analysis, pol sequences appeared to be less variable than env and LTR sequences. In addition, we performed a Shannon entropy-based per-site variability profile of the pol , env and LTR sequences used in this study (Fig. 2 A-C).
Sequence variability measured as per-site entropy. A Multiple alignment of the pol gene showing the locations of qPCR fragments in regions of the pol gene for the qPCR1 (highlighted in pink), qPCR4 (highlighted in yellow) and for the qPCR7, qPCR8, qPCR9, qPCR10 and qPCR11 assays (highlighted in orange). B Multiple alignment of the env gene targeted by ddPCR6 (highlighted by blue rectangle). C Multiple alignment of the LTR region by qPCR3 (highlighted in mint)
The all-observed entropy plots were homogeneous along the whole sequences. Considering the three regions of pol gene, the highest entropy (4.67) occurred in the region targeted by the qPCR1 primers, whereas the entropy for qPCR7—qPCR11 and qPCR4 primers were 1.57 and 0.38, respectively. For the LTR region targeted by qPCR3 primers and for env gene targeted by ddPCR6, the total entropy was equal to 4.46 and 7.85, respectively. This analysis showed a marked region of variability for LTR and env fragments. Interestingly, we noted that the qPCR7—qPCR11 targeted the most conserved regions of reverse transcriptase and qPCR4 primers targeted the most-conserved region of virus integrase (Fig. 2 A-C; see also Additional file 7).
Quantitation of BLV proviral DNA by different qPCR/ddPCR assays
To analyze whether the range of copy numbers detected by each qPCR was comparable to those of the others, Kruskal–Wallis one-way analysis of variance (ANOVA) was used. The violin plots were used to visualize the ANOVA results (Fig. 3 A-B).
Comparison of detection of BLV proviral DNA copy numbers by eleven testing methods. Shown is a box plot of data from Kruskal–Wallis ANOVA, a rank test. The DNA copy numbers for 41 samples, determined independently by each of the 11 qPCRs, were used for the variance analysis. In this analysis, the positive controls (sample ID 42 and ID 43) and negative control (sample ID 32) were excluded. A Violin plot for graphical presentation of the ANOVA of proviral copy number values. B Violin plot for ANOVA analysis of variance, copy number values are presented on a logarithmic scale (Log1.2) for better illustration of copy number differences between PCR methods
The grouping variable revealed significant differences among the distributions of proviral DNA copy numbers with the various qPCRs ( P < 0.001). These results showed that the abilities of qPCRs/ddPCR to determine the proviral DNA copy number differed. A Dunn-Bonferroni test was used to compare the groups in pairs to find out which was significantly different. The Dunn-Bonferroni test revealed that the pairwise group comparisons of qPCR2—qPCR4, qPCR3—ddPCR6, qPCR4—qPCR5, qPCR4—ddPCR6, qPCR4—qPCR9, qPCR4—qPCR10, qPCR5—qPCR11, ddPCR6—qPCR11 and qPCR9—qPCR11 have an adjusted P value less than 0.05 and thus, it can be assumed that these groups were significantly different in each pair (see Additional file 4). The Pareto chart was used to show the average copy number values of all methods in descending order. These Pareto charts were prepared based on 80–20 rule, which states that 80% of effects come from 20% of the various causes [ 46 ]. The methods that generated the highest copy numbers was qPCR3 and qPCR4, on the other hand the lowest copy numbers and/or highest negative results were generated by ddPCR6 (Fig. 4 ).
A Pareto chart with the proviral BLV copy mean values for eleven PCR assay arranged in descending order. Pareto charts was prepared based on 80–20 rule, which states that 80% of effects come from 20% of the various causes
The correlations between copy numbers detected by different qPCRs and ddPCR assays were calculated. The Kendall's Tau correlation coefficient measured between each pair of the assays was shown in the Additional file 5 and in Fig. 5 as a correlation heatmap. The average correlation for all qPCRs and ddPCR assays was strong (Kendall's tau = 0.748; P < 0.001).
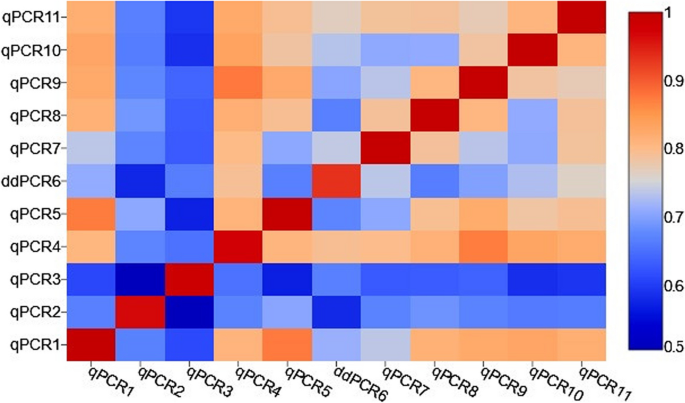
The heatmap of Kendall’s tau correlation coefficients between copy numbers detected by ten qPCRs and one ddPCR. Statistically significant differences in the distribution of copy numbers, a moderate, strong and very strong correlation between particular qPCRs/ddPCR was observed. The strength of the association, for absolute values of r, 0–0.19 is regarded as very weak, 0.2–0.39 as weak, 0.40–0.59 as moderate, 0.6–0.79 as strong and 0.8–1 as very strong correlation
Since the differences between PCR tests may be influenced by the number of BLV proviral copies present in each sample, we compared the average number of BLV copies between a group of genomic DNA samples that gave concordant results (group I [ n = 28]) and a group that gave discordant results (group II [ n = 15]). The mean number of copies was 73,907 (minimum, 0; maximum, 4,286,730) in group I, and 3,479 (minimum, 0; maximum, 218,583) in group II, and this difference was statistically significant ( P < 0.001 by a Mann–Whitney U- test) (Fig. 6 ).
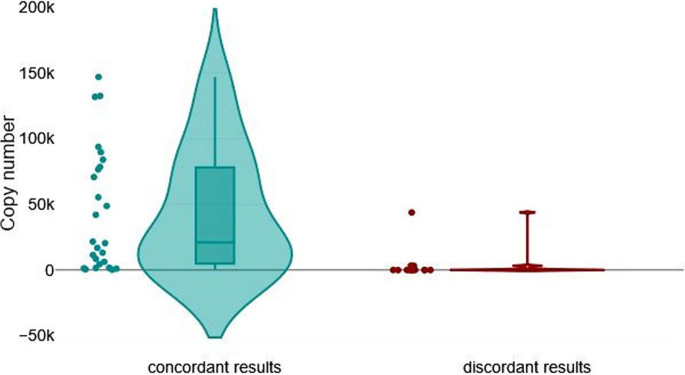
Impact of BLV proviral copy numbers on the level of agreement. Violin plot for graphical presentation of Mann–Whitney U test. The test was performed to compare BLV provirus copy number in two groups of samples: 28 samples with fully concordant results from all eleven qPCR/ddPCR assays (left) and 15 samples with discordant results from different qPCR/ddPCR assays (right) ( P < 0.001). Sample ID 42 was excluded from the statistical analysis
The results show that the concordant results group had considerably higher copy numbers (median, 5,549.0) than the discordant results group (median, 6.3).
BLV control and eradication programs consist of correct identification and subsequent segregation/elimination of BLV-infected animals [ 47 ]. Detection of BLV- infected cows by testing for BLV-specific antibodies in serum by agar gel immunodiffusion and ELISA is the key step and standard to be implemented of EBL eradication programs according to WOAH ( https://www.woah.org/en/disease/enzootic-bovine-leukosis/) [ 9 ]. Despite the low cost and high throughput of serological tests, there are several scenarios where highly specific and sensitive molecular assays for the detection of BLV DNA might improve detection and program efficiency.
In this perspective, qPCR assays can detect small quantities of proviral DNA during acute infection, in which animals show very low levels of anti-BLV antibodies [ 43 , 48 , 49 , 50 ]. qPCR methods can also work as confirmatory tests to clarify ambiguous and inconsistent serological test results [ 11 ]. Such quantitative features of qPCRs are crucial when eradication programs progress and prevalence decreases. Moreover, qPCR allows not only the detection of BLV infection but also estimation of the BLV PVL, which directly correlates with the risk of disease transmission [ 51 , 52 ]. This feature of qPCR allows for a rational segregation of animals based on the stratified risk of transmission. These considerations allow for greater precision in the management of BLV within large herds with a high prevalence of BLV ELISA-positive animals to effectively reduce herd prevalence [ 13 , 53 ]. BLV is a global burden and the lack of technical standardization of molecular detection systems remains a huge obstacle to compare surveillance data globally based on the first interlaboratory trial performed in 2018 [ 15 ]. In the 2018 study we observed an adjusted level of agreement of 70% comparing qualitative qPCR results; however, inconsistencies amongst methods were larger when low number of copies of BLV DNA were compared. Samples with low copies of BLV DNA (< 20 copies per 100 ng) accounted for the higher variability and discrepancies amongst tests. We concluded from the first interlaboratory trial that standardizing protocols to improve sensitivity of assays with lower detection rates was necessary.
In this follow up study, we re-tested the TaqMan BLV qPCR developed and validated by NVRI (acting as reference WOAH laboratory) and the one adapted from this original protocol to be used with SYBR Green dye, allowing a significant reduction in costs [ 11 ]. Another 3 laboratories also performed NVRI´s qPCR with slight modifications (i.e., Spain performed a multiplex assay for internal normalization). The remaining 6 labs introduced novel methodologies to the trial including one ddPCR (UY).
To compare different qPCR methods, a more comprehensive sample panel, accounting for a more geographical diversification was used in this trial. The amounts of BLV DNA in these samples were representative of the different BLV proviral loads found in field samples (from 1 to > 10,000 copies of BLV proviral DNA). Of note, 34% of reference samples had less than 100 copies of BLV DNA per 100 ng; samples were lyophilized to grant better preservation and reduced variability during distribution to participants around the globe.
The panel included a single negative control and two positive controls. Diagnostic sensitivity (DxSn) was estimated for each qPCR. Considering the 43 positive samples, the DxSn for the different qPCRs were: qPCR1 = 100%, qPCR2 = 82%, qPCR3 = 86%, qPCR4 = 84%, qPCR5 = 93%, ddPCR6 = 74%, qPCR7 = 93%, qPCR8 = 98%, qPCR9 = 98%, qPCR10 = 98% and qPCR11 = 95%. The most sensitive method was the qPCR1, and the method with the lowest sensitivity was the ddPCR6 method. Twenty-nine out of 44 samples were identified correctly by all qPCRs. The remaining 15 samples gave discordant results. The comparison of qualitative qPCR results among all raters revealed an overall observed agreement of 87%, indicating strong interrater reliability (Cohen´s kappa = 0.396) [ 54 , 55 ].
There are several factors that contribute to variability in qPCR results (i.e., number of copies of target input, sample acquisition, processing, storage and shipping, DNA purification, target selection, assay design, calibrator, data analysis, etc.). For that reason and as expected, the level of agreement among sister qPCRs (qPCR7, qPCR9-11) sharing similar protocols was higher compared to the rest of assays; this was also true for qPCR8 which targets the same region of BLV pol gene (shares same primers) but has a particular set-up to be used with SYBR Green chemistry. Oppositely, lower sensitivity and larger discrepancy against other tests was observed for the ddPCR6 and qPCR2-4.
Based on these observations we investigated which factors might have accounted for larger assessment variability amongst tests. In the first place, we observed that the use of different chemistries was not detrimental for the sensitivity and agreement among tests; similar DxSn and comparable level of agreement were obtained comparing TaqMan (qPCR7, 10, 11) vs SYBR Green (qPCR8) chemistries while targeting identical BLV sequence and using same standards. Also, when a multiplex qPCR (TaqMan) targeting the same BLV sequence and using the same standard was compared to previous ones, agreement was kept high, indicating that the lower sensitivity described for some multiplex qPCRs did not take place in this comparison. The use of an international calibrator and the efficiency estimation (standard curve) might inform variability associated with different chemistries. In contrast, another multiplex assay targeting another region of BLV pol (qPCR2) showed much lower sensitivity and agreement. As qPCR2 is performed as service by private company and oligonucleotide sequences were not available, we were not able to investigate in which proportion each of these two variables contributed to the lower performance of this assay, but we note the addition of 4 µl genomic DNA to this assay that would have an impact the DxSn. In this regard, there is substantial evidence showing that the variability of target sequence among strains from different geographical areas, might affect the sensitivity of BLV qPCRs. Previous studies comparing the pol , gag , tax and env genes reported that the pol gene was the most suitable region to target for diagnostic purposes, since it provided the most-sensitive assays [ 11 , 15 , 56 , 57 , 58 , 59 ]. This might be due in part to higher sequence conservation of pol among strains from different geographical areas. Supporting this observation, it is noticeable how JPN qPCR improved their performance in the current trial, by targeting pol in place of tax , as it did in the previous interlaboratory trial. Since it is a commercial test, we cannot exclude other factors contributing for the performance upgrade observed for this qPCR. In the current study, qPCR3 and ddPCR6 targeting LTR and env sequences, showed lower performances than other assays. Standardization of DNA input into each qPCR would have likely resulted in higher concordance in results. For instance, qPCR1 added 10 µl of genomic DNA per reaction and ddPCR6 added 1 µl of genomic DNA, impacting the resulting sensitivity differences.
Since the sensitivity of each assay and, consequently, the level of agreement among assays might also be influenced by the number of BLV DNA copies present in each sample [ 48 ], we compared the average number of BLV DNA copies between a group of genomic DNA samples that gave concordant results and a group that gave discordant results, and observed that samples that gave discordant results had significantly lower numbers of BLV DNA copies than samples that gave concordant results. Related to this point, the degradation of target DNA during lyophilization, shipment and resuspension, could have been more significant in low-copy compared to high-copy samples. Consequently, the degradation of target DNA in samples with low copies of BLV DNA might have accounted for the greater level of discrepancy within this subset of samples. The rational of adding a large proportion of such samples (34% samples with less than 100 BLV copies per 100 ng of total DNA) was to mimic what is frequently observed in surveillance programs (i.e., hyperacute infection, chronic asymptomatic infection, etc.).
Quantitative methods for the detection of BLV DNA copies are important for segregation programs based on animal level of BLV PVL, as well as for scientific research and the study of BLV dynamics. When the numbers of copies of BLV DNA detected by different assays were compared, in the present study, we observed that although the ability to quantify BLV DNA differed among qPCRs/ddPCR and there were statistically significant differences in the distribution of copy numbers among assays, a strong average correlation was found for the eleven qPCRs/ddPCR. In this regard, the lack of an international calibrator (standard curve) could be a major contributor to the increment of quantitative variation amongst laboratories. For that reason, plasmid pBLV1 containing pol 120 bp sequence was originally constructed for use as standard for quantification and shared with some collaborators (i.e., qPCR7, qPCR8, qPCR 9, qPCR10 and qPCR11). Remarkably, the laboratories used pBLV1 standard in the current trial obtained the most comparable results, indicating that the use of an international standard may have significant impact on the convergence of results; such standard reference material should be prepared under identical conditions. To avoid further variability a detailed protocol for lyophilized DNA sample resuspension, quantitation and template input into each qPCR should be shared with all participants.
Conclusions
BLV DNA was detected with different level of sensitivity in serologically positive samples from different origin and classified into different BLV genotypes. Overall agreement was high; however, we found significant differences in results for the samples with low BLV DNA copy numbers. This second interlaboratory study demonstrated that differences in target sequence, DNA input and calibration curve standards can increase interlaboratory variability considerably. Next steps should focus on (i) standard unification (international gold standard) to estimate individual test efficiency and improve quantitative accuracy amongst tests; (ii) building a new panel of samples with low BLV DNA copy numbers to re-evaluate sensitivity and quantitation of molecular methods. Since no variation was observed in samples from different genotypes, all samples will be collected in Poland to standardize the collection, purification, lyophilization and shipping steps with precise instructions for suspension and constant input volume for the PCR reaction. Finally, we believe that following this standardization approach we will be able to improve overall agreement amongst tests, improving the diagnostic of BLV around the world.
Availability of data and materials
Not applicable.
Data availability
No datasets were generated or analysed during the current study.
Abbreviations
One-way analysis of variance
Bovine leukemia virus
BLV-specific antibodies
Digital PCR
Diagnostic sensitivity
Enzootic bovine leukosis
Enzyme-linked immunosorbent assays
Real-time fluorescence resonance energy transfer PCR
Genomic quality number
Histone H3 family 3A housekeeping gene
Maximum likelihood phylogenetic tree
Multiple-sequence alignment
Peripheral blood leukocytes
Phosphate-buffered saline
Proviral load
Quantitative real-time PCR
Room temperature
World Organisation for Animal Health
Coffin JM, Hughes SH, Varmus HE. (Eds.). 1650–1655 (1997). Retroviruses. Cold Spring Harbor Laboratory Press.
Ghysdael J, Bruck C, Kettmann R, Burny A. Bovine leukemia virus. Curr Top Microbiol Immunol. 1984;112:1–19.
CAS PubMed Google Scholar
Ott SL, Johnson R, Wells SJ. Association between bovine-leukosis virus seroprevalence and herd-level productivity on US dairy farms. Prev Vet Med. 2003;61:249–62.
Article CAS PubMed Google Scholar
Bartlett PC, et al. Options for the control of bovine leukemia virus in dairy cattle. J Am Vet Med Assoc. 2014;244:914–22.
Article PubMed Google Scholar
Kuczewski A, et al. Economic evaluation of 4 bovine leukemia virus control strategies for Alberta dairy farms. J Dairy Sci. 2019;102:2578–92.
Frie MC, Coussens PM. Bovine leukemia virus: a major silent threat to proper immune responses in cattle. Vet Immunol Immunopathol. 2015;163:103–14.
Panel, E.A. Scientific opinion on enzootic bovine leukosis. EFSA J. 2015;13:4188.
Google Scholar
OIE. World Animal Health Information Database - Version: 1.4. World Animal Health Information Database. Paris, France: World Organisation for Animal Health; 2009. Available from: http://www.oie.int . Accessed 16 Aug 2024.
Health, W.O.f.A. Manual of diagnostic tests and vaccines for terrestrial animals. Infect Bursal Dis. 2012;12:549–65.
Hutchinson HC, et al. Bovine leukemia virus detection and dynamics following experimental inoculation. Res Vet Sci. 2020;133:269–75.]
Rola-Luszczak M, Finnegan C, Olech M, Choudhury B, Kuzmak J. Development of an improved real time PCR for the detection of bovine leukaemia provirus nucleic acid and its use in the clarification of inconclusive serological test results. J Virol Methods. 2013;189:258–64.
Nakada S, Kohara J, Makita K. Estimation of circulating bovine leukemia virus levels using conventional blood cell counts. J Dairy Sci. 2018;101:11229–36.
Ruggiero VJ, Bartlett PC. Control of Bovine Leukemia Virus in Three US Dairy Herds by Culling ELISA-Positive Cows. Vet Med Int. 2019;2019:3202184.
Article PubMed PubMed Central Google Scholar
Kobayashi T, et al. Increasing Bovine leukemia virus (BLV) proviral load is a risk factor for progression of Enzootic bovine leucosis: A prospective study in Japan. Prev Vet Med. 2020;178: 104680.
Article Google Scholar
Jaworski JP, Pluta A, Rola-Łuszczak M, McGowan SL, Finnegan C, Heenemann K, Carignano HA, Alvarez I, Murakami K, Willems L, Vahlenkamp TW, Trono KG, Choudhury, B, Kuźmak J. Interlaboratory Comparison of Six Real-Time PCR Assays for Detection of Bovine Leukemia Virus Proviral DNA. J Clin Microbiol. 2018;56(7):e00304-18. https://doi.org/10.1128/JCM.00304-18 .
Pluta A, Rola-Luszczak M, Douville RN, Kuzmak J. Bovine leukemia virus long terminal repeat variability: identification of single nucleotide polymorphisms in regulatory sequences. Virol J. 2018;15:165.
Article CAS PubMed PubMed Central Google Scholar
Pluta A, Willems L, Douville RN, Kuźmak J. Effects of Naturally Occurring Mutations in Bovine Leukemia Virus 5'-LTR and Tax Gene on Viral Transcriptional Activity. Pathog. 2020;9(10):836. https://doi.org/10.3390/pathogens9100836 .
Pluta A, et al. Molecular characterization of bovine leukemia virus from Moldovan dairy cattle. Arch Virol. 2017;162:1563–76.
Rola-Łuszczak M, Sakhawat A, Pluta A, Ryło A, Bomba A, Bibi N, Kuźmak J. Molecular Characterization of the env Gene of Bovine Leukemia Virus in Cattle from Pakistan with NGS-Based Evidence of Virus Heterogeneity. Pathogens (Basel, Switzerland). 2021;10(7):910. https://doi.org/10.3390/pathogens10070910 .
Rola-Luszczak M, et al. The molecular characterization of bovine leukaemia virus isolates from Eastern Europe and Siberia and its impact on phylogeny. PLoS ONE. 2013;8: e58705.
Pinheiro de Oliveira TF, et al. Detection of contaminants in cell cultures, sera and trypsin. Biologicals. 2013;41:407–14.
Pluta A, Blazhko NV, Ngirande C, Joris T, Willems L, Kuźmak J. Analysis of Nucleotide Sequence of Tax, miRNA and LTR of Bovine Leukemia Virus in Cattle with Different Levels of Persistent Lymphocytosis in Russia. Pathogens. 2021;10(2):246. https://doi.org/10.3390/pathogens10020246 .
Yang Y, et al. Bovine leukemia virus infection in cattle of China: Association with reduced milk production and increased somatic cell score. J Dairy Sci. 2016;99:3688–97.
DeGraves FJ, Gao D, Kaltenboeck B. High-sensitivity quantitative PCR platform. Biotechniques. 2003;34(106–110):112–105.
Fonseca Junior AA, et al. Evaluation of three different genomic regions for detection of bovine leukemia virus by real-time PCR. Braz J Microbiol. 2021;52:2483–8.
Farias MVN, et al. Toll-like receptors, IFN-gamma and IL-12 expression in bovine leukemia virus-infected animals with low or high proviral load. Res Vet Sci. 2016;107:190–5.
Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5’––3’ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A. 1991;88:7276–80.
De Brun ML, et al. Development of a droplet digital PCR assay for quantification of the proviral load of bovine leukemia virus. J Vet Diagn Invest. 2022;34:439–47.
Rola-Łuszczak M, Finnegan C, Olech M, Choudhury B, Kuźmak J. Development of an improved real time PCR for the detection of bovine leukaemia provirus nucleic acid and its use in the clarification of inconclusive serological test results. J Virol Methods. 2013;189:258–64.
Petersen MI, Alvarez I, Trono KG, Jaworski JP. Quantification of bovine leukemia virus proviral DNA using a low-cost real-time polymerase chain reaction. J Dairy Sci. 2018;101:6366–74.
Toussaint JF, Sailleau C, Breard E, Zientara S, De Clercq K. Bluetongue virus detection by two real-time RT-qPCRs targeting two different genomic segments. J Virol Methods. 2007;140:115–23.
John EE, et al. Development of a predictive model for bovine leukemia virus proviral load. J Vet Intern Med. 2022;36:1827–36.
Farias MVN, et al. Toll-like receptors, IFN-γ and IL-12 expression in bovine leukemia virus-infected animals with low or high proviral load. Res Vet Sci. 2016;107:190–5.
Yoneyama S, et al. Comparative Evaluation of Three Commercial Quantitative Real-Time PCRs Used in Japan for Bovine Leukemia Virus. Viruses. 2022;14:1182.
Polat M, Takeshima SN, Aida Y. Epidemiology and genetic diversity of bovine leukemia virus. Virol J. 2017;14:209.
Lee E, et al. Molecular epidemiological and serological studies of bovine leukemia virus (BLV) infection in Thailand cattle. Infect Genet Evol. 2016;41:245–54.
Duran-Yelken S, Alkan F. Molecular analysis of the env, LTR, and pX regions of bovine leukemia virus in dairy cattle of Türkiye. Virus Genes. 2024;60:173–85.
Lv G, Wang J, Lian S, Wang H, Wu R. The Global Epidemiology of Bovine Leukemia Virus: Current Trends and Future Implications. Animals. 2024;14(2):297. https://doi.org/10.3390/ani14020297 .
Úsuga-Monroy C, Díaz FJ, Echeverri-Zuluaga JJ, González-Herrera LG, López-Herrera A. Presence of bovine leukemia virus genotypes 1 and 3 in Antioquia, Colombia. Revista UDCA Actualidad & Divulgación Científica. 2018;21:119–26.
Úsuga-Monroy C, Díaz FJ, González-Herrera LG, Echeverry-Zuluaga JJ, López-Herrera A. Phylogenetic analysis of the partial sequences of the env and tax BLV genes reveals the presence of genotypes 1 and 3 in dairy herds of Antioquia. Colombia VirusDisease. 2023;34:483–97.
Martin D, et al. Comparative study of PCR as a direct assay and ELISA and AGID as indirect assays for the detection of bovine leukaemia virus. J Vet Med B Infect Dis Vet Public Health. 2001;48:97–106.
Cohen J. A Coefficient of Agreement for Nominal Scales. Educ Psychol Measur. 1960;20:37–46.
Feinstein AR, Cicchetti DV. High agreement but low kappa: I. The problems of two paradoxes. J Clin Epidemiol. 1990;43:543–9.
Woolhouse MEJ, et al. Heterogeneities in the transmission of infectious agents: Implications for the design of control programs. Proc Natl Acad Sci. 1997;94:338–42.
Ohshima K, Okada K, Numakunai S, Kayano H, Goto T. An eradication program without economic loss in a herd infected with bovine leukemia virus (BLV). Nihon Juigaku Zasshi. 1988;50:1074–8.
Juliarena MA, Gutierrez SE, Ceriani C. Determination of proviral load in bovine leukemia virus-infected cattle with and without lymphocytosis. Am J Vet Res. 2007;68:1220–5.
Mirsky ML, Olmstead CA, Da Y, Lewin HA. The prevalence of proviral bovine leukemia virus in peripheral blood mononuclear cells at two subclinical stages of infection. J Virol. 1996;70:2178–83.
Eaves FW, Molloy JB, Dimmock CK, Eaves LE. A field evaluation of the polymerase chain reaction procedure for the detection of bovine leukaemia virus proviral DNA in cattle. Vet Microbiol. 1994;39:313–21.
Juliarena MA, Barrios CN, Ceriani MC, Esteban EN. Hot topic: Bovine leukemia virus (BLV)-infected cows with low proviral load are not a source of infection for BLV-free cattle. J Dairy Sci. 2016;99:4586–9.
Yuan Y, et al. Detection of the BLV provirus from nasal secretion and saliva samples using BLV-CoCoMo-qPCR-2: Comparison with blood samples from the same cattle. Virus Res. 2015;210:248–54.
Taxis TM, et al. Reducing bovine leukemia virus prevalence on a large midwestern dairy farm by using lymphocyte counts, ELISA antibody testing, and proviral load. The Bovine Practitioner. 2020;54:136–44.
McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb). 2012;22:276–82.
Cicchetti DV, Feinstein AR. High agreement but low kappa: II. Resolving the paradoxes J Clin Epidemiol. 1990;43:551–8.
Heenemann K, et al. Development of a Bovine leukemia virus polymerase gene–based real-time polymerase chain reaction and comparison with an envelope gene–based assay. J Vet Diagn Invest. 2012;24:649–55.
Lew AE, et al. Sensitive and specific detection of proviral bovine leukemia virus by 5′ Taq nuclease PCR using a 3′ minor groove binder fluorogenic probe. J Virol Methods. 2004;115:167–75.
Takeshima SN, Sasaki S, Meripet P, Sugimoto Y, Aida Y. Single nucleotide polymorphisms in the bovine MHC region of Japanese Black cattle are associated with bovine leukemia virus proviral load. Retrovirology. 2017;14:24.
Debacq C, et al. Reduced proviral loads during primo-infection of sheep by Bovine Leukemia virus attenuated mutants. Retrovirology. 2004;1:31.
Kuckleburg CJ, et al. Detection of bovine leukemia virus in blood and milk by nested and real-time polymerase chain reactions. J Vet Diagn Invest. 2003;15:72–6.
Dube S, et al. Degenerate and specific PCR assays for the detection of bovine leukaemia virus and primate T cell leukaemia/lymphoma virus pol DNA and RNA: phylogenetic comparisons of amplified sequences from cattle and primates from around the world. J Gen Virol. 1997;78(Pt 6):1389–98.
Download references
Acknowledgements
The authors thank Luc Willems (University of Liège, Belgium) for plasmid DNA sample pBLV344; Marlena Smagacz and Eliza Czarnecka (National Veterinary Research Institute, Poland) for lyophilizing DNA samples and DNA analysis, respectively; Ali Sakhawat (Animal Quarantine Department, Pakistan), Vitaliy Bolotin (National Scientific Center IECVM, Ukraine), Frank van der Meer and Sulav Shrestha (University of Calgary, Canada) for sharing material.
The APC was funded by the National Veterinary Research Institute, Puławy, Poland.
Author information
Authors and affiliations.
Department of Biochemistry, National Veterinary Research Institute, Puławy, 24-100, Poland
Aneta Pluta & Jacek Kuźmak
Instituto de Virología E Innovaciones Tecnológicas (IVIT), Centro de Investigaciones en Ciencias Veterinarias y Agronómicas (CICVyA), Instituto Nacional de Tecnología Agropecuaria (INTA) - CONICET, Buenos Aires, Argentina
Juan Pablo Jaworski & Vanesa Ruiz
CentralStar Cooperative, 4200 Forest Rd, Lansing, MI, 48910, USA
Casey Droscha & Sophie VanderWeele
Department of Animal Science, College of Agriculture and Natural Resources, Michigan State University, East Lansing, Michigan, 48824, USA
Tasia M. Taxis
Niort Laboratory, Unit Pathology and Welfare of Ruminants, French Agency for Food, Environmental and Occupational Health and Safety (Anses), Ploufragan-Plouzané, Niort, France
Stephen Valas
Croatian Veterinary Institute, Savska Cesta 143, Zagreb, 10000, Croatia
Dragan Brnić & Andreja Jungić
Laboratorio Central de Veterinaria (LCV), Ministry of Agriculture, Fisheries and Food, Carretera M-106 (Km 1,4), Madrid, Algete, 28110, Spain
María José Ruano & Azucena Sánchez
Department of Veterinary Sciences, Faculty of Agriculture, Iwate University, 3-18-8 Ueda, Morioka, 020-8550, Japan
Kenji Murakami & Kurumi Nakamura
Departamento de Patobiología, Facultad de Veterinaria, Unidad de Microbiología, Universidad de La República, Ruta 8, Km 18, Montevideo, 13000, Uruguay
Rodrigo Puentes & MLaureana De Brun
Laboratorio de Virología, Departamento SAMP, Centro de Investigación Veterinaria de Tandil-CIVETAN (CONICET/UNCPBA/CICPBA), Buenos Aires, Argentina
Marla Eliana Ladera Gómez, Pamela Lendez & Guillermina Dolcini
Laboratório Federal de Defesa Agropecuária de Minas Gerais, Pedro Leopoldo, Brazil
Marcelo Fernandes Camargos & Antônio Fonseca
Department of Pathobiology, College of Veterinary Medicine, Auburn University, Auburn, AL, 36849-5519, USA
Subarna Barua & Chengming Wang
Department of Omics Analyses, National Veterinary Research Institute, 24-100, Puławy, Poland
Aneta Pluta & Aleksandra Giza
You can also search for this author in PubMed Google Scholar
Contributions
Proposed the conception and design of the study, A.P.; data curation, A.P., J.P.J., C.D., S.V., D.B., A.S., K.M., R.P., G.D., M.F.C. and CH.W.; investigation, A.P., V.R., S.VW., S.V., A.J., M.J.R., K.N., M.L.B., M.L.G., P.L., A.F., A.G. and S.B., formal analysis, A.P.; statistical analysis, A.P.; database analysis, A.P., visualization of the results, A.P.; resources, A.P., T.M.T. and J.K; writing—original draft preparation, A.P., J.P.J.; writing—review and editing, A.P., J.P.J., C.D., S.VW., T.M.T. and J.K; project administration, A.P. All authors read and approved the submitted version.
Corresponding author
Correspondence to Aneta Pluta .
Ethics declarations
Ethics approval and consent to participate.
The study was approved by the Veterinary Sciences Animal Care Committee No. AC21-0210, Canada; the Institutional Animal Care and Use Committee No. PROTO202000096 from 4/13/2020 to 4/14/2023, Michigan State University, United States and the Ethics Review Board, COMSATS Institute of Information Technology, Islamabad, Pakistan, no. CIIT/Bio/ERB/17/26. Blood samples from Polish, Moldovan and Ukrainian cattle, naturally infected with BLV, were selected from collections at local diagnostic laboratories as part of the Enzootic bovine leukosis (EBL) monitoring program between 2012 and 2018 and sent to the National Veterinary Research Institute (NVRI) in Pulawy for confirmation study. The approval for collection of these samples from ethics committee was not required according to Polish regulation (“Act on the Protection of Animals Used for Scientific or Educational Purposes”, Journal of Laws of 2015). All methods were carried out in accordance with relevant guidelines and regulations. The owners of the cattle herds from which the DNA samples originated, the district veterinarians caring for these farms and the ministries of agriculture were informed and consented to the collection of blood from the animals for scientific purposes and the sending of samples to NVRI.
Consent for publication
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
12917_2024_4228_moesm1_esm.pdf.
Additional file 1. Copy of the instruction included with the panel of 44 DNA samples sent to participating laboratories for dilution of the lyophilisates
12917_2024_4228_MOESM2_ESM.png
Additional file 2. Detection of the H3F3A gene copy number in 43 DNA samples; no outlier was found for any samples ( P <0.05) (two-sided).
12917_2024_4228_MOESM3_ESM.docx
Additional file 3. Concentration values of 44 DNA samples measured by the 11 participating laboratories (given in ng per µl)
12917_2024_4228_MOESM4_ESM.pdf
Additional file 4. Post hoc - Dunn-Bonferroni-Tests. The Dunn-Bonferroni test revealed that the pairwise group comparisons of qPCR2 - qPCR4, qPCR3 - ddPCR6, qPCR4 - qPCR5, qPCR4 - ddPCR6, qPCR4 - qPCR9, qPCR4 - qPCR10, qPCR5 - qPCR11, ddPCR6 - qPCR11 and qPCR9 - qPCR11 have an adjusted p-value less than 0,05
12917_2024_4228_MOESM5_ESM.docx
Additional file 5. Kendall's Tau correlation coefficient values measured between each pair of assays. The numbers 1 to 11 in the first column and last row of the table indicate the names of the assays qPCR1-qPCR5, ddPCR6, qPCR7-qPCR11 respectively
12917_2024_4228_MOESM6_ESM.png
Additional file 6. Maximum-likelihood phylogenetic analysis of full-length BLV-pol gene sequences representing 7 BLV genotypes (G1, G2, G3, G4, G6, G9, and G10) (A); (B) env-based sequences assigned to 10 BLV genotypes (G1, G2, G3, G4, G5, G6, G7, G8, G9, and G10); (C) LTR-based sequences representing 10 BLV genotypes (G1-G10). For all genes and LTR region the Tamura-Nei model and Bootstrap replications (1,000) were applied in MEGA X
12917_2024_4228_MOESM7_ESM.pdf
Additional file 7. Multiple sequence alignment of reverse transcriptase, integrase, envelope and LTR sequences in the context of the specific primers used by different qPCR assays. (A) Multiple sequence alignment of reverse transcriptase (pol gene) sequences in the context of qPCR7, qPCR8, qPCR9, qPCR10 and qPCR11 assay primers. (B) Multiple sequence alignment of integrase (pol gene) sequences in the context of qPCR4 assay primers. (C) Multiple sequence alignment of env gene sequences in the context of ddPCR6. (D) Sequence alignment of LTR region sequences in the context of qPCR3 method primers
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Pluta, A., Jaworski, J.P., Droscha, C. et al. Inter-laboratory comparison of eleven quantitative or digital PCR assays for detection of proviral bovine leukemia virus in blood samples. BMC Vet Res 20 , 381 (2024). https://doi.org/10.1186/s12917-024-04228-z
Download citation
Received : 24 November 2023
Accepted : 09 August 2024
Published : 26 August 2024
DOI : https://doi.org/10.1186/s12917-024-04228-z
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Bovine leukemia virus ( BLV)
- Quantitative real-time PCR (qPCR)
- Proviral DNA
- BLV international network
- Update on the efforts in harmonization qPCR
BMC Veterinary Research
ISSN: 1746-6148
- General enquiries: [email protected]
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 28 August 2024
Gut Microbiome Wellness Index 2 enhances health status prediction from gut microbiome taxonomic profiles
- Daniel Chang ORCID: orcid.org/0000-0003-0076-2833 1 na1 ,
- Vinod K. Gupta 2 na1 ,
- Benjamin Hur 2 ,
- Sergio Cobo-López 3 ,
- Kevin Y. Cunningham 4 ,
- Nam Soo Han 5 ,
- Insuk Lee ORCID: orcid.org/0000-0003-3146-6180 6 ,
- Vanessa L. Kronzer ORCID: orcid.org/0000-0002-7489-3134 7 ,
- Levi M. Teigen ORCID: orcid.org/0000-0002-6409-0485 8 ,
- Lioudmila V. Karnatovskaia 9 ,
- Erin E. Longbrake 10 ,
- John M. Davis III 7 ,
- Heidi Nelson 11 &
- Jaeyun Sung ORCID: orcid.org/0000-0002-3005-2798 2 , 7 , 12
Nature Communications volume 15 , Article number: 7447 ( 2024 ) Cite this article
5 Altmetric
Metrics details
- Applied microbiology
- Predictive markers
- Predictive medicine
Recent advancements in translational gut microbiome research have revealed its crucial role in shaping predictive healthcare applications. Herein, we introduce the Gut Microbiome Wellness Index 2 (GMWI2), an enhanced version of our original GMWI prototype, designed as a standardized disease-agnostic health status indicator based on gut microbiome taxonomic profiles. Our analysis involves pooling existing 8069 stool shotgun metagenomes from 54 published studies across a global demographic landscape (spanning 26 countries and six continents) to identify gut taxonomic signals linked to disease presence or absence. GMWI2 achieves a cross-validation balanced accuracy of 80% in distinguishing healthy (no disease) from non-healthy (diseased) individuals and surpasses 90% accuracy for samples with higher confidence (i.e., outside the “reject option”). This performance exceeds that of the original GMWI model and traditional species-level α-diversity indices, indicating a more robust gut microbiome signature for differentiating between healthy and non-healthy phenotypes across multiple diseases. When assessed through inter-study validation and external validation cohorts, GMWI2 maintains an average accuracy of nearly 75%. Furthermore, by reevaluating previously published datasets, GMWI2 offers new insights into the effects of diet, antibiotic exposure, and fecal microbiota transplantation on gut health. Available as an open-source command-line tool, GMWI2 represents a timely, pivotal resource for evaluating health using an individual’s unique gut microbial composition.
Similar content being viewed by others

A predictive index for health status using species-level gut microbiome profiling

Taxonomic signatures of cause-specific mortality risk in human gut microbiome
Multi-omic integration of microbiome data for identifying disease-associated modules
Introduction.
Recent landmark studies have unveiled profound links between the gut microbiome and a variety of complex, chronic diseases 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 . Despite these discoveries, how can we tell if a person has dysbiosis? How can we effectively harness unique microbial signatures to quantitatively track our health? These critical questions stand at the forefront of utilizing the gut microbiome as a precise marker for health and wellness.
The potential of the gut microbiome as a marker for deciphering complex, chronic diseases has captivated the scientific community—in response, we recently developed the Gut Microbiome Wellness Index (GMWI) [previously called the Gut Microbiome Health Index (GMHI)] 10 . GMWI is a first-of-its-kind stool metagenome-based indicator for assessing health by determining the likelihood of an individual harboring a clinically diagnosed disease solely from their gut microbiome composition, irrespective of the specific disease type 10 , 11 . This disease-agnostic index was derived from a comprehensive analysis of a pooled dataset comprising 4347 stool shotgun metagenomes from 34 independent studies. GMWI is a logarithmic ratio of the collective abundances—a term encompassing species-level relative abundances and multiple α-diversity metrics—of health- and disease-associated gut microbial species. Evaluating on the pooled dataset, GMWI exhibited a balanced accuracy (i.e., average of the proportions of healthy and non-healthy samples that were correctly classified) of 69.7% in predicting the presence of clinically diagnosed disease. Specifically, the correct classification rates for healthy (disease-free) individuals and those with non-healthy (diseased) conditions were 75.6% and 63.8%, respectively. Moreover, GMWI achieved a balanced accuracy of 73.7% in a validation cohort of 679 stool metagenomes, with the correct classification rates for the healthy and non-healthy subsets being 77.1% (91 out of 118) and 70.2% (394 out of 561), respectively. Since its original publication in 2020, GMWI has been utilized in studies investigating the impact of environmental 12 and genetic/socioeconomic 13 factors on the human gut microbiome, as well as in identifying a ‘Longevous Gut Microbiota Signature’ species set 13 .
Despite the promise of our original GMWI prototype, there are limitations that impede its general applicability. Firstly, GMWI correctly classifies healthy stool metagenomes at a higher success rate than non-healthy ones. This bias may stem from the prevalence-based strategy used to identify health-associated and disease-associated species, which was a fundamental component of the GMWI model. As the non-healthy group encompasses patients with different diseases, this group is inherently heterogeneous; in turn, a prevalence-based strategy may miss subtle taxonomic signatures that are only represented in subsets of non-healthy populations (e.g., cohorts with a specific disease). Secondly, our existing model assigns equal weight to each species without considering potential variances in the importance of individual species. To improve classification accuracy and general applicability, a refined weighting system that accounts for varying strengths of association to host phenotype is needed. Additionally, including gut microbial information from all taxonomic ranks could uncover more features that accurately predict host phenotypes 14 , 15 . In this study, we present GMWI2, an advanced iteration of the original GMWI that addresses the above limitations and significantly improves classification accuracy in distinguishing between healthy and non-healthy phenotypes.
Pooled analysis of stool metagenomes across health and disease phenotypes
As in our previous work 10 , we define “healthy” subjects as those without reported diseases or abnormal body weight conditions (i.e., classified as underweight, overweight, or obese based on reported BMI), whereas “non-healthy” subjects are those confirmed to have a clinical diagnosis of any disease. (Retaining the same definitions for “healthy” and “non-healthy” ensures that the current work represents a continuous refinement of our original GMWI method.) We conducted a pooled analysis of existing 8069 stool shotgun metagenomes (5547 from healthy individuals and 2522 from non-healthy individuals) sourced from 54 independently published studies spanning 26 countries and six continents (Fig. 1a , Table 1 , and Supplementary Data 1 ). These pooled metagenomes are from individuals with one of twelve different health and disease phenotypes (Fig. 1a ; healthy, ankylosing spondylitis, atherosclerotic cardiovascular disease, colorectal cancer, Crohn’s disease, Graves’ disease, liver cirrhosis, multiple sclerosis, nonalcoholic fatty liver disease (or also known as metabolic dysfunction-associated steatotic liver disease [MASLD]), rheumatoid arthritis, type 2 diabetes, and ulcerative colitis) from diverse geographies, ethnicities/races, cultures, and balanced sex representation (Fig. 1b ). (Our study and sample selection criteria can be found in the “Methods” section. We provide all subjects’ phenotype, age, sex, BMI, and geography [as provided in their respective original study] in Supplementary Data 2 .) This substantial increase in sample size, nearly doubling the number of metagenomes included in our previous study, is one notable improvement in GMWI2. Additionally, GMWI2 uses MetaPhlAn3 16 instead of MetaPhlAn2 17 for taxonomic profiling, leveraging an extensively expanded marker database for a more comprehensive and accurate characterization of microbial taxa (“Methods” section).
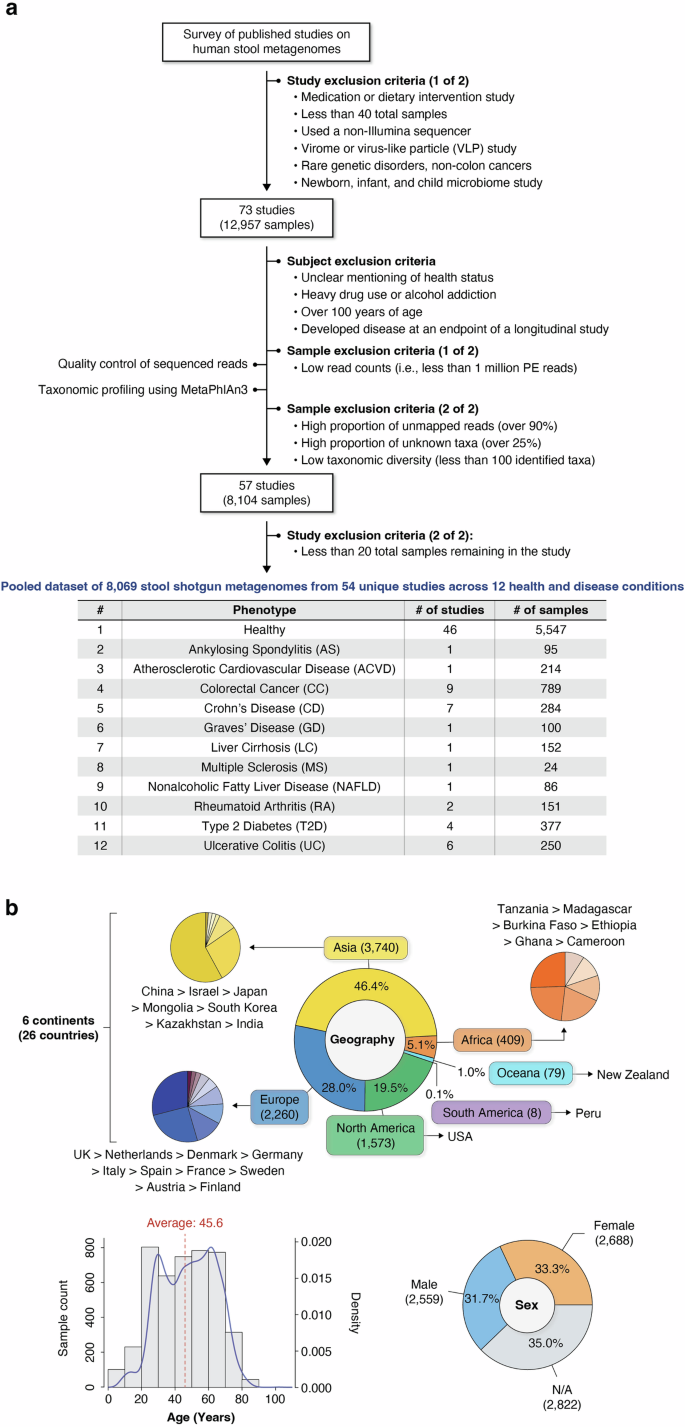
a A survey was conducted in PubMed and Google Scholar to search for published studies with publicly available human stool shotgun metagenome (gut microbiome) samples from healthy (disease-free) and non-healthy (diseased) individuals. The initial collection of stool metagenomes consisted of 12957 samples from 73 independent studies. All raw metagenome samples (.fastq files) were downloaded and reprocessed uniformly using identical bioinformatics methods. After quality control of sequenced reads, taxonomic profiling was performed using MetaPhlAn3. Studies and samples were removed based on several exclusion criteria. Finally, a total of 8069 samples (5547 and 2522 metagenomes from healthy and non-healthy individuals, respectively) from 54 studies ranging across healthy and 11 non-healthy phenotypes were assembled into a pooled metagenome dataset for downstream analyses. b Demographic summary of the study subjects whose metagenome samples were included in the pooled dataset. Subject demographics, as reported in the original studies, include country of origin ( n = 8069), age ( n = 4670), and sex ( n = 5247).
All metagenomes underwent uniform reprocessing using an identical bioinformatics pipeline, as described in the “Methods” section. Such practice not only mitigates batch effects 18 , 19 , but also bolsters the identification of health- and disease-related gut taxonomic signatures despite the presence of potentially strong confounding factors. Indeed, this is supported by principal component analysis (PCA), where, despite the samples originating from varying sources and conditions, the healthy and non-healthy groups display significantly distinct gut microbiome profiles (Adonis R 2 = 1.2%, P = 0.001, PERMANOVA; Fig. 2a ). Nevertheless, although the consensus preprocessing of metagenomic data effectively reduces one source of batch effects related to bioinformatics analyses, it is important to recognize that this approach cannot entirely eliminate potential batch effects arising from experimental and technical procedures across different studies. Such factors include differences in how stool samples were collected, stored, and prepared for metagenomic sequencing.
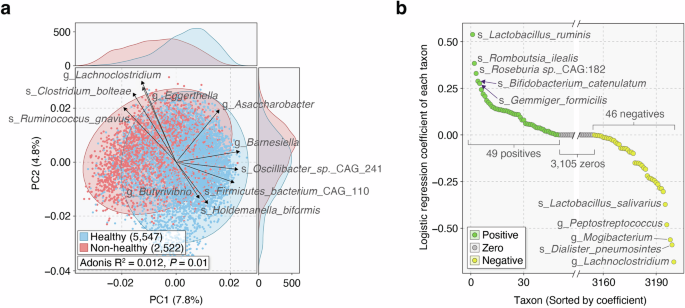
a Principal component analysis (PCA) of gut microbiome profiles. Significant differences in distributions between healthy (disease-free) (blue, n = 5547) and non-healthy (diseased) (red, n = 2522) groups were observed ( P < 0.05, PERMANOVA). Ellipses represent 95% confidence regions. The loading vectors with the top 10 highest PC1 and PC2 magnitudes are shown. b Coefficient values for the Lasso-penalized logistic regression model. The model includes 49 taxa with positive coefficients, 3105 taxa with zero coefficients, and 46 taxa with negative coefficients.
Implementing Lasso-penalized logistic regression in GMWI2
For the classification task of distinguishing between healthy and non-healthy groups, GMWI2 uses a Lasso-penalized logistic regression model instead of the log-ratio equation utilized in the original GMWI. Hence, GMWI2 essentially uses linear regression for its predictions, resembling polygenetic risk score models in statistical genetics 20 , 21 . The model was trained on gut microbiome taxonomic profiles (derived from the aforementioned pooled dataset of 8069 stool shotgun metagenomes) spanning all measurable taxonomic ranks to model disease likelihood as a linear function of microbial taxon (i.e., clade) presence or absence. Specifically, the GMWI2 score for an individual sample is defined as the predicted log odds (logit) of the sample originating from a healthy, non-diseased individual. A more comprehensive explanation of how GMWI2 uses Lasso-penalized logistic regression to estimate disease likelihood is detailed in “Methods” section.
The original GMWI approach utilized a prevalence-based strategy to identify health- and disease-associated microbial species. Our current method learns variable feature importances, obviating the need for manual species identification. More specifically, the Lasso-penalized logistic regression model utilized 95 microbial taxa with non-zero coefficients for its predictions, derived directly from the gut microbiome profiles (Fig. 2b and Supplementary Data 3 ). Interestingly, the majority of taxa characterized by positive and negative coefficients exhibited a higher relative abundance in the healthy and non-healthy groups, respectively (Supplementary Data 4 ). These identified taxa included 1 class, 3 orders, 4 families, 19 genera, and 68 species. Notably, the coefficient values varied between –0.68 and 0.54, ensuring that each taxon contributes differently to the GMWI2 score according to its relative association strength. This presents a shift from our previous GMWI log-ratio model where equal weight was assigned to each species.
It is worth mentioning that several taxonomic levels exhibited non-zero coefficients in our analysis. This is likely due in part to the interdependence across different levels of taxonomic hierarchy introducing multicollinearity, which complicates the interpretation of regression coefficients. However, our approach in encompassing all taxonomic levels demonstrated higher classification performance compared to when using only a single taxonomic level (Supplementary Table 1 ). Given our primary objective of optimizing classification accuracy, we chose to prioritize this aspect, leading us to set aside the multicollinearity concern.
In the following sections, we evaluate GMWI2’s proficiency in differentiating healthy from non-healthy individuals. This process can be conceptually structured into four phases:
Model training: GMWI2 is trained and evaluated on the full training dataset. This phase utilizes all 8069 samples for computing the logistic regression coefficients (as depicted in Fig. 2b ) and determining GMWI2 scores.
Cross-validation: GMWI2 undergoes further evaluation through cross-validation (CV) and inter-study validation (ISV) strategies. In contrast to the initial phase, these strategies do not leverage all 8069 samples simultaneously for model training. As a result, the models generated during this phase are intrinsically different from those produced in the first phase. In line with standard cross-validation protocols, the training of the GMWI2 model, including the computation of logistic regression coefficients, is confined strictly to the training partition of each train-test split of the total 8069 samples.
Validation on external datasets: The GMWI2 model developed in the first phase is applied to six external datasets to confirm its discriminatory power on independent samples.
Demonstration on longitudinal datasets: The GMWI2 model from the first phase is applied to four additional external datasets. These evaluations focus on demonstrating GMWI2’s applicability in longitudinal scenarios.
Enhanced classification of healthy and non-healthy gut microbiomes with GMWI2
GMWI2 scores were calculated for metagenomes by applying the learned coefficients in computing the predicted log odds. A positive GMWI2 value classifies the sample as healthy, indicating disease absence; while a negative GMWI2 value classifies it as non-healthy, denoting disease presence. A GMWI2 of 0 implies an equal weighted presence of positive coefficient taxa and negative coefficient taxa, thereby classifying the sample as neither healthy nor non-healthy. When evaluated on the training dataset (8069 samples), GMWI2 demonstrated a balanced accuracy of 79.9% (correct classification rate in healthy: 79.2%, correct classification rate in non-healthy: 80.6%) and a Cliff’s Delta ( d ) effect size of 0.75, significantly surpassing the balanced accuracy and Cliff’s Delta reported by our original GMWI model (71.8%, d = 0.63) and traditional species-level α-diversity indices (i.e., Shannon Index, Simpson Index, and richness) (Fig. 3a and Supplementary Data 5 ). Our results indicate that GMWI2 differentiates between healthy and non-healthy groups much more effectively than GMWI, although both indices were strongly correlated (Pearson’s r = 0.81; Supplementary Fig. 1 ). Moreover, we found that the gut microbiomes of healthy individuals exhibit significantly higher GMWI2 scores compared to each of the eleven disease phenotypes (Fig. 3b ). Lastly, we observed weak correlations between GMWI2 and clinical/demographic characteristics ( | Spearman’s ρ | < 0.3; Supplementary Figs. 2a–g ), such as age, BMI, fasting blood glucose, blood cholesterol and triglycerides, indicating that these factors do not significantly influence gut microbiome-based classification outcomes.
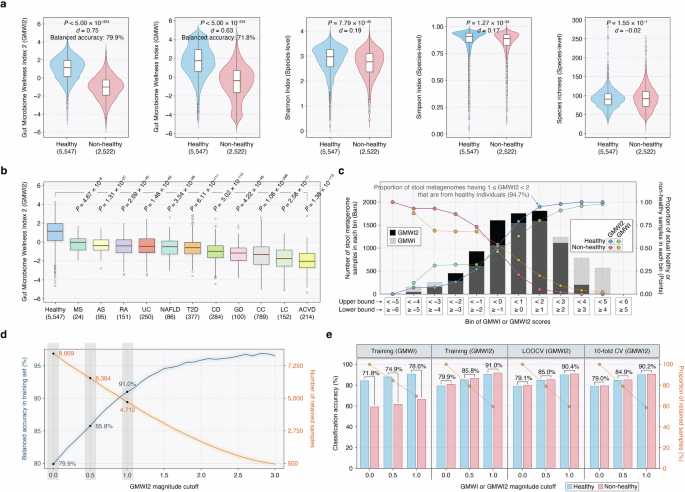
a GMWI2 best stratifies healthy ( n = 5547) and non-healthy ( n = 2522) groups compared to GMWI and α-diversity indices ( P -values from the two-sided Mann–Whitney U test; d , Cliff’s Delta effect size). Balanced accuracies on the training set are shown for GMWI2 and GMWI. b The healthy group (blue, far left) exhibits significantly higher GMWI2 scores than all 11 non-healthy phenotypes ( P -values from the two-sided Mann–Whitney U test). Non-healthy phenotypes include multiple sclerosis (MS, n = 24), ankylosing spondylitis (AS, n = 95), rheumatoid arthritis (RA, n = 151), ulcerative colitis (UC, n = 250), nonalcoholic fatty liver disease (NAFLD, n = 86), type 2 diabetes (T2D, n = 377), Crohn’s disease (CD, n = 284), Graves’ disease (GD, n = 100), colorectal cancer (CC, n = 789), liver cirrhosis (LC, n = 152), and atherosclerotic cardiovascular disease (ACVD, n = 214). c Bins of GMWI2 and GMWI scores (x-axis). The height of the black and gray bars indicate metagenome sample counts in each GMWI2 and GMWI bin, respectively (y-axis, left). Points represent the proportion of samples in each GMWI2 or GMWI bin corresponding to actual healthy and non-healthy individuals (y-axis, right). d Increased magnitude cutoffs result in improved classification performance of GMWI2, showing increasing training set balanced accuracy (blue, y-axis, left) at the expense of decreasing retained samples (orange, y-axis, right). e Classification performances of GMWI and GMWI2 in distinguishing healthy and non-healthy groups. Accuracies (y-axis, left) are depicted for both groups on the training set, leave-one-out cross-validation (LOOCV), and 10-fold CV, using varying magnitude cutoffs (0, 0.5, 1.0) of GMWI and GMWI2 scores. Balanced accuracies are shown between the blue and pink bars, which represent healthy and non-healthy groups, respectively. Orange points represent the proportion of retained samples (y-axis, right) for the corresponding index magnitude cutoff. For 10-fold CV, repeated random sub-sampling was performed ten times, and the average results are displayed. Standard box-and-whisker plots (i.e., center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, outliers) are used to depict groups of numerical data in ( a , b ).
We subsequently explored whether higher (or more positive) GMWI2 values could indicate enhanced confidence in categorizing stool metagenomes as healthy. Conversely, we examined if lower (or more negative) GMWI2 scores suggest an increased likelihood that a sample could be classified as non-healthy. Indeed, we observed a progressive increase in the proportion of healthy individuals among metagenome samples with increasingly positive GMWI2 scores (Fig. 3c and Supplementary Table 2 ). Similarly, increasingly negative GMWI2 scores captured larger proportions of the non-healthy subjects. Notably, the proportions of actual healthy and non-healthy samples within the positive and negative bins of GMWI2, respectively, were both higher compared to the same GMWI bins (refer to points in Fig. 3c ). This difference in sample distributions between the GMWI2 and GMWI bins underscores GMWI2’s improved capability to differentiate between healthy and non-healthy samples.
The results presented in Fig. 3c of our study revealed an interesting trend. Specifically, when GMWI2 (and GMWI) scores exhibit a more positive or negative value, there is a corresponding increase in the proportion of actual healthy and non-healthy samples, respectively. This trend suggests a potential increase in the confidence of phenotype classification. In contrast, as these values near zero, our confidence in accurately determining the presence or absence of a disease decreases. To examine this point more closely, we next investigated how setting a minimum GMWI2 threshold or cutoff parameter could enhance classification accuracy for phenotype prediction. We observed remarkable improvement in classification performance when considering increasing cutoffs for the magnitude of GMWI2 scores, thereby signifying higher prediction confidence in the retained samples (Supplementary Table 3 ). For example, when retaining samples with GMWI2 magnitudes equal to or higher than 0.5 (i.e., GMWI2 scores below –0.5 or above +0.5) and 1.0 (i.e., GMWI2 scores below –1.0 or above +1.0), we achieved balanced accuracies of 85.8% and 91.0%, respectively (Fig. 3d ). (these cutoffs are examples to illustrate the concept of the GMWI2 magnitude cutoff.) This approach, however, requires excluding samples with GMWI2 magnitudes below these cutoffs, leaving only 6364 (representing 78.9% of the total 8069 samples) and 4712 (58.4% of 8069) samples, respectively. This highlights a significant trade-off: increasing the cutoff improves accuracy but excludes potentially valuable samples from the analysis.
An important observation is that GMWI2 correctly classified healthy and non-healthy stool metagenomes at nearly the same rate (79.2% and 80.6%, respectively) despite imbalanced sample numbers. This contrasts markedly with the original GMWI, which achieved a much higher correct classification rate on healthy samples (Fig. 3e ). We also assessed the performance of the GMWI2 model utilizing both leave-one-out cross-validation (LOOCV) and 10-fold cross-validation (10-fold CV) (Fig. 3e ). Interestingly, GMWI2 achieved nearly identical balanced accuracies of 79.1% (healthy correct classification rate: 78.6%, non-healthy correct classification rate: 79.5%) and 79.0% (healthy correct classification rate: 78.6%, non-healthy correct classification rate: 79.3%) in LOOCV and 10-fold CV, respectively, nearly matching the performance achieved on the training dataset (79.9%).
Next, we computed classification accuracies using different magnitude cutoffs for the two cross-validation methods (Fig. 3e ). Remarkably, GMWI2 achieved a balanced accuracy of 90.4% and 90.2% in LOOCV and 10-fold CV, respectively, on the samples with scores below –1.0 or above +1.0. These balanced accuracies were very close to those observed in the training set (91.0%). In contrast, when applying the same criteria to GMWI (i.e., cutoff of 1.0), the balanced accuracy drops considerably to 78.6%. In all, these results emphasize the notable improvements achieved with GMWI2 over GMWI.
Evaluating the robustness of GMWI2 across study populations of varying sample sizes
Although studies with small sample sizes were excluded from the training set (see study exclusion criteria in Fig. 1a and “Methods” section), in general, it is crucial to validate any classification model on datasets of varying sample sizes 19 . To this end, we conducted inter-study validation (ISV) to assess the impact of batch effects (i.e., technical or biological variations associated with the study population or site characteristics) on GMWI2 performance stability. In this approach, we iteratively excluded a single study, trained the GMWI2 model on the remaining studies, and evaluated its classification performance on the held-out study 22 . (The excluded study essentially becomes the independent validation [or test] cohort.) An important aspect of ISV is that it can showcase the significant variability in classification performance that can arise depending on the choice of validation set. For our study, it provides a range of classification accuracies achievable when applying GMWI2 across 54 independent validation sets.
Figure 4a specifically displays the performance of GMWI2 across the full range of held-out studies, along with details on their sample sizes. Despite the variation in classification performance across different studies (see gold points indicating ISV classification accuracy per study in Fig. 4a and Supplementary Table 4 ), the average balanced accuracy was 75.8%. This performance rose to 86.9% when considering samples with GMWI2 scores lower than –1 or higher than 1 (Supplementary Table 4 ). In all, our analysis revealed no discernible correlation between the model’s predictive performance and the sample size of the held-out datasets.
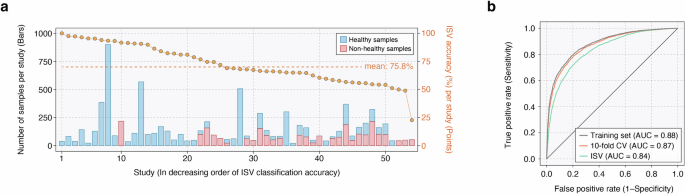
a Classification accuracy on each excluded study in ISV is displayed by gold points (y-axis, right). The studies on the x-axis are rank-ordered based on either accuracy for a single phenotype (healthy or non-healthy) or balanced accuracy in the case of both phenotypes. The stacked bars illustrate the number of healthy (blue) and non-healthy (pink) stool metagenome samples in each study (y-axis, left). b Receiver operating characteristic curves for classification performance in distinguishing healthy and non-healthy phenotypes on the training set, 10-fold CV, and ISV.
The classification performances obtained from ISV exhibited minimal disparity compared to the performances achieved by LOOCV and 10-fold CV, which do not consider study boundaries. The small discrepancy between these strategies shows GMWI2’s resilience against batch-related biases, indicating that GMWI2 generalizes effectively across stool metagenomes, regardless of the subjects’ origins. Further evidence of this robustness is demonstrated by the area-under-the-curve (AUC) metrics in the training set, 10-fold CV, and ISV, achieving AUCs of 0.88, 0.87, and 0.84, respectively (Fig. 4b ).
Demonstration of GMWI2 predictive capability on independent sample sets
To confirm GMWI2’s predictive capability for distinguishing between healthy and non-healthy individuals, we compiled an external validation dataset consisting of 1140 stool metagenome samples from six published studies (Supplementary Data 6 ). This dataset includes samples from healthy individuals and patients diagnosed with ankylosing spondylitis, pancreatic cancer, or Parkinson’s disease. All metagenome samples in this validation dataset (Supplementary Data 7 ) were classified into either healthy or non-healthy groups in the same manner as demonstrated above.
Consistent with our findings from the discovery cohort (or training data), GMWI2 scores from stool metagenomes of the healthy validation group ( n = 494) were significantly higher than those of the non-healthy validation group ( n = 646) ( P = 1.6 × 10 –43 , two-sided Mann–Whitney U test; Cliff’s Delta = 0.48; Fig. 5a ). The balanced accuracy achieved was 72.1%, which is comparable to the average balanced accuracy of 75.8% observed in our ISV analysis. With magnitude cutoffs of 0.5 and 1.0, the balanced accuracy improved to 75.4% and 80.1%, respectively, while still retaining 74.3% and 49.3% of the samples.
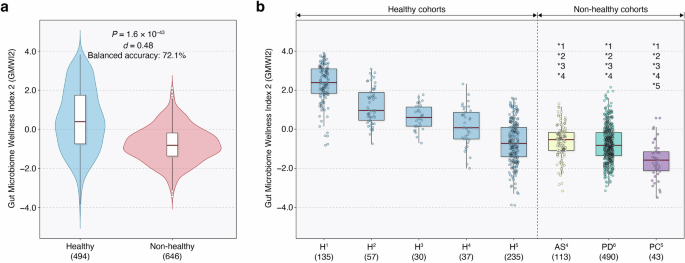
a GMWI2 scores from healthy (494 samples) and non-healthy (646 samples) groups. Scores are significantly higher in the healthy group compared to the non-healthy group ( P = 1.6 × 10 –43 ; two-sided Mann–Whitney U test). The effect size is represented by Cliff’s Delta ( d = 0.48). The balanced accuracy of the classification is 72.1%. b GMWI2 scores across five healthy (H 1 –H 5 ) and three non-healthy cohorts (AS 4 ankylosing spondylitis, PD 6 Parkinson’s disease, PC 5 pancreatic cancer). The superscript numbers adjacent to phenotype abbreviations correspond to specific studies detailed in Supplementary Data 6 . Asterisk (*) indicates significantly higher score in a healthy cohort compared to the corresponding non-healthy cohort ( P < 0.01, two-sided Mann–Whitney U test. Exact P -values provided in Supplementary Data 6 ). Numbers next to each asterisk refer to the healthy cohort compared against each non-healthy condition. Sample size of each group or cohort are shown in parentheses. Standard box-and-whisker plots (i.e., center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, outliers in ( a ) or individual GMWI2 scores in ( b )) are used to depict groups of numerical data.
To further examine GMWI2 performance on the external validation data, we analyzed the eight total cohorts (defined by unique phenotype per study), spanning five healthy and three non-healthy phenotypes. As shown in Fig. 5b , four of the five healthy cohorts (H 1 –H 4 ) were found to have significantly higher GMWI2 distributions than all three non-healthy phenotype cohorts ( P < 0.01, two-sided Mann–Whitney U test). Classification accuracies for the five healthy cohorts were as follows: 96.3% (130 of 135) for H 1 , 91.2% (52 of 57) for H 2 , 83.3% (25 of 30) for H 3 , 56.8% (21 of 37) for H 4 , and 28.1% (66 of 235) for H 5 . Alternatively, classification accuracies for the three non-healthy cohorts were 90.7% (39 of 43) for pancreatic cancer (PC 5 ), 81.2% (398 of 490) for Parkinson’s disease (PD 6 ), and 80.5% (91 of 113) for ankylosing spondylitis (AS 4 ). Notably, GMWI2 performed well (81.2%) in predicting adverse health in Parkinson’s disease, although stool metagenomes from patients with this neurodegenerative disorder were not part of the original discovery set. Furthermore, despite the relatively poor classification performance in the H 5 cohort (28.1%), the GMWI2 scores in H 5 were significantly higher than those in the PC 5 pancreatic cancer group from the same study. Overall, the robust reproducibility of GMWI2 on an external validation dataset suggests that a generalized disease-associated signature of gut microbiome dysbiosis across multiple diseases was effectively captured during dataset integration and index formulation.
Gut health tracking in longitudinal studies
We applied GMWI2 to stool metagenomes obtained from four recently published longitudinal gut microbiome studies. Importantly, these samples were not part of the initial pool of 8069 metagenomes used to train GMWI2. Here, our aim was to illustrate GMWI2’s versatility by demonstrating it towards gut microbiome health tracking, thereby extending its applicability beyond the originally intended case vs. control scenarios. Our index for quantitatively monitoring gut health can be likened to using a cholesterol and glucose test for evaluating cardiovascular and metabolic health over time.
Using data from the first study 23 , we analyzed stool metagenomes from 22 individuals with irritable bowel syndrome (IBS) before and six months after receiving fecal microbiota transplantation (FMT) from two healthy donors. Among the participants, 14 reported symptom relief after FMT (“Effect” group), while 8 did not experience symptom relief (“No Effect” group) despite both groups demonstrating a significant increase in species richness at six months following FMT ( P < 0.05, one-sided Wilcoxon signed-rank test; Supplementary Fig. 3 ). However, only the individuals in the “Effect” group exhibited a significant increase in GMWI2 ( P < 0.05; Fig. 6a and Supplementary Table 5 ). Likewise, an increase in the species-level Shannon Index was observed only in the “Effect” group ( P < 0.05; Supplementary Fig. 4 ). Overall, these findings suggest that while α-diversity metrics, such as richness and Shannon diversity, may yield conflicting conclusions, changes in GMWI2 could serve as a marker of subjects’ phenotypes following FMT treatment for IBS. Furthermore, in light of the clinical significance and the complexities involved in donor screening for FMT 24 , 25 , computational tools such as GMWI2 (given its more nuanced definition of gut health) may be able to help guide the selection of suitable healthy donors and their stool samples.
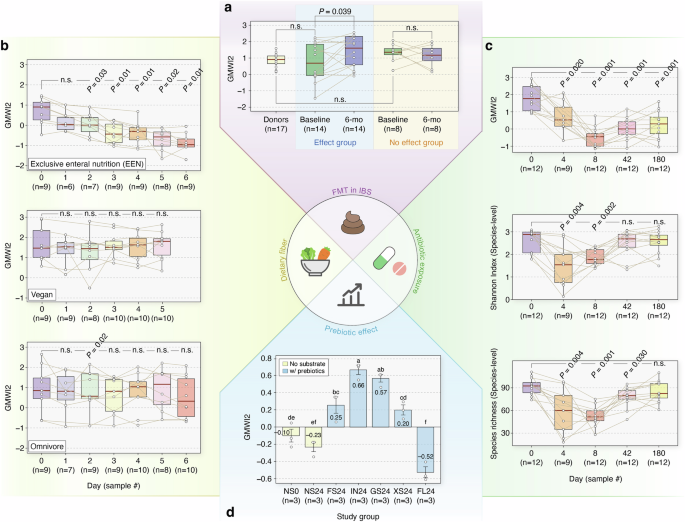
a Changes in GMWI2 in patients with irritable bowel syndrome observed six months (6-mo) after undergoing fecal microbiota transplantation. Only subjects experiencing symptom relief (“Effect” group) displayed a significant increase in GMWI2 ( P = 0.039, one-sided Wilcoxon signed-rank test). n , number of FMT donor samples (17 total samples from two healthy donors) or number of FMT recipients. b GMWI2 scores for dietary groups (EEN, Vegan, and Omnivore) at baseline and at the first 5–6 days of dietary intervention. The EEN group showed significant changes in GMWI2, with values significantly decreased by day 2 and thereafter ( P < 0.05, two-sided Wilcoxon signed-rank test). No significant change in GMWI2 was observed for the Omnivore and Vegan groups compared to baseline. n, number of unique individuals who each provided a stool sample per time point. c GMWI2, Shannon Index, and species richness before and after antibiotic intervention. Despite recovery in Shannon Index and species richness at day 42 and day 180, respectively, GMWI2 remained significantly lower compared to day 0, suggesting incomplete gut microbiome recovery even after ~6 months ( P < 0.05, two-sided Wilcoxon signed-rank test). n , number of unique individuals who each provided a stool sample per time point. d GMWI2 of gut microbial communities after 24-h in vitro fecal fermentation with five different prebiotic oligosaccharides. The experiment was conducted in triplicates for each study group. The height of the bars represents the mean GMWI2 (numbers inside the solid bars), and error bars indicate the standard deviation from the mean. Points represent individual triplicate samples. Different small letters above the bars denote groups with significant differences in GMWI2 as determined by Tukey’s HSD test ( P < 0.05). Control groups: NS0, no substrate addition at 0 h; NS24, no substrate for 24 h. Prebiotic groups: FS24 fructooligosaccharide, IN24 inulin, GS24 galactooligosaccharide, XS24 xylooligosaccharide, FL24 2’-fucosyllactose. Standard box-and-whisker plots (i.e., center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, individual GMWI2 scores or α-diversity values) are used to depict groups of numerical data in ( a – c ).
In the second study 26 , we investigated the effects of diet. We calculated GMWI2 for stool metagenomes obtained from 30 healthy volunteers before and during a dietary intervention. Three groups of participants were studied: Vegan (self-reported vegans who resumed their regular diet), Omnivore (participants who consumed a standard diet of both animal and plant origin), and Exclusive Enteral Nutrition (EEN) (participants with an omnivorous diet who went on to consume a synthetic, fiber-free diet for the duration of the study). Stool samples were collected at baseline and each day during the dietary intervention. We observed that the GMWI2 scores for both the vegan and omnivore subjects remained relatively stable throughout the intervention period of five to six days (Fig. 6b ). However, GMWI2 for the EEN group significantly decreased relative to baseline by the second day and onwards ( P < 0.05, two-sided Wilcoxon signed-rank test; Fig. 6b and Supplementary Table 6 ) while α-diversities did not significantly change across the groups (Supplementary Fig. 5 ). These results suggest that the removal of dietary fiber may lead to a rapid decrease in overall gut health, an early change detected solely by GMWI2 and not by α-diversity metrics. Overall, our findings strengthen the evidence for the well-established benefits of dietary fiber on health 27 , 28 , 29 .
For the third study 30 , we calculated GMWI2 for stool metagenomes from twelve healthy young adults who underwent a 4-day exposure with broad-spectrum antibiotics (meropenem, gentamicin, and vancomycin). Here, stool samples were collected before the exposure, and then again at 4, 8, 42, and 180 days post-intervention. While species-level α-diversity measures (Shannon Index and richness) indicated that the gut microbiome may have recovered somewhat by day 42 or 180, GMWI2 did not demonstrate any recovery trend even by day 180 (Fig. 6c and Supplementary Table 7 ). These findings reflect deleterious post-intervention taxonomic shifts originally noted by Palleja et al., such as the rise in previously undetectable Clostridium spp ., and the disappearance of probiotic members of Bifidobacterium and butyrate producers Coprococcus eutactus and Eubacterium ventriosum . Our results therefore offer a novel perspective on the long-term impact of short-term broad-spectrum antibiotic intervention on gut microbiota and suggest that GMWI2 could be a valuable tool for assessing gut microbiome recovery following an acute illness.
In the final study 31 , we examined the effect of various oligosaccharides on gut microbial communities. In this study, Lee et al. used GMWI to assess the prebiotic effect of oligosaccharides, with broader implications for designing personalized diets based on their impact on gut microbiome wellness. Herein, 19 healthy adult volunteers (14 men and 5 women) provided fecal samples, which were then combined and well-mixed. Then, fructooligosaccharides (FOS), galactooligosaccharides (GOS), xylooligosaccharides (XOS), inulin (IN), and 2′-fucosyllactose (2FL) were separately mixed with portions of the homogenized fecal samples in a 24-h in vitro anaerobic batch fecal fermentation system. Two control groups were also included: one without substrate addition at 0 h (NS0) and another without substrate addition for 24 h (NS24). The experiment was conducted in triplicates for each of the seven study groups.
GMWI2 was calculated for all fecal samples (Fig. 6d and Supplementary Table 8 ), thereby replicating the original study with our new index. Consistent with previous findings, the NS24 group exhibited a lower average GMWI2 than the NS0 group, indicating a less healthy and more disease-associated state. Notably, the addition of the three prebiotics (FOS, IN, and GOS) resulted in significantly higher GMWI2 compared to NS0 ( P < 0.05, Tukey’s HSD test). Also, these same three prebiotics, along with XOS, led to significantly higher GMWI2 relative to NS24 ( P < 0.05). However, unlike the GMWI2 results, traditional α-diversity metrics (Shannon Index, species richness, species evenness, and inverse Simpson’s Index) were reported to have significantly lower values in all prebiotic treatment groups compared to the NS0 group ( P < 0.05) 31 . Therefore, at least in the in vitro fermentation setting, intake of these four prebiotics could potentially stimulate the growth of gut microbial species associated with healthy conditions, an effect observed solely by using GMWI2.
Recent research into the human gut microbiome has highlighted its potential to inform the development of innovative tools for predictive healthcare 32 , 33 , 34 , 35 , 36 , 37 . In this regard, we introduce GMWI2, a robust predictor of health status based on gut microbiome taxonomic profiles that display significant technological advances compared to its prototype (GMWI). Our extensive multi-study analysis, pooling 8069 stool shotgun metagenomes from 54 published studies, encompasses a diverse range of demographics from 26 countries across six continents to identify the biological signals linking gut taxonomies to human health. Delivering a cross-validation balanced accuracy of approximately 90% for higher confidence samples, GMWI2 establishes its strong reliability as a classifier that distinguishes between healthy and non-healthy phenotypes. Furthermore, by revisiting and reinterpreting data from previously published datasets, GMWI2 can offer novel perspectives even for the established understanding of the impact of dietary influences, antibiotic exposure, and FMT on the gut microbiome. Lastly, this study highlights the importance of extensive data sharing in fostering robust machine learning applications, and in demonstrating resilience to batch effects and biases 22 , 38 , 39 , 40 .
In our analyses in which we incrementally increased the GMWI2 magnitude cutoff, we recognize an inverse relationship between classification accuracy and the volume of samples eligible for class prediction. Therefore, constraining this magnitude cutoff to a single value may not be universally applicable; instead, the selection of this parameter should be flexible and determined by the user, tailored to the specific context and acceptable accuracy thresholds of their individual datasets. In other words, users can select their desired GMWI2 magnitude cutoff based on their confidence level preference in the predictions. This user-driven approach, which offers flexibility between high confidence in a limited dataset and broader range predictions with lesser confidence, is a distinct advantage of our method over traditional binary-output machine learning techniques. Moreover, our findings thus foster the potential utility of a “reject option” 41 , 42 for low GMWI2 magnitudes, which can serve as a criterion to redirect relatively uncertain predictions to other screening methods—this concept captures the understanding that certain aspects of health and disease are not fully explainable solely by the gut microbiome.
Our study, while providing insights into the predictive capabilities of the gut microbiome, has some limitations that need to be acknowledged. First and foremost, we emphasize that GMWI2 scores reflect an association with health status, which we define in terms of the presence or absence of disease. It is important to understand that these scores do not imply a causal relationship with (nor are they intended to replace) direct clinical health measures, such as the detection of pathogenic organisms in the gastrointestinal tract, gut motility characteristics, metabolic profiles, serological markers, blood inflammatory markers, or fecal calprotectin levels. Second, the model could benefit from the inclusion of more intricate microbiome features such as species growth rates, strain details, and functional potential. Incorporating these important factors may improve predictive accuracy and offer a richer perspective on the intricate mechanisms tying the gut microbiome to overall human health. Third, we made concerted efforts to ensure that our pooled stool metagenomic dataset exhibits a diverse representation of geographies, races, and cultures. Nevertheless, future work should emphasize wider participant inclusion, especially from underrepresented areas and ethnicities, to truly globalize gut microbiome research. Additionally, loosening our selection criteria will allow us to incorporate metagenomes from a broader range of disease phenotypes (like neurodegenerative and psychiatric disorders) and reach even more diverse demographics. Such expansion could enhance the model’s generalizability across different populations. Fourth, although we utilized taxonomic information down to the species level, there’s a potential missed opportunity in not focusing on microbial strains, which often bear more clinical significance. While our method surpasses the genus-level limitations of 16S rRNA gene amplicon sequencing, it doesn’t account for the variability among strains of the same species. Fifth, our analysis revealed that well-known pathogens, including Enterococcus faecium/faecalis , did not display negative coefficients in our GMWI2 framework. Nevertheless, we did observe negative coefficients for certain opportunistic pathogenic taxa, notably among various Clostridium species, as detailed in Supplementary Data 4 . It is important to emphasize that the determination of pathogenic traits is more accurately conducted at the strain level, which falls outside the scope of our model. Additionally, it is widely acknowledged that not every gut microbiome associated with chronic, non-communicable diseases necessarily harbors invasive pathogens. Sixth, we recognize that the compositional shifts between healthy and non-healthy identified by our model might be influenced by variables such as transit time, stool consistency, and other factors not captured in our meta-data. This is a valid consideration for individual samples. However, in our analysis of over 8000 metagenome samples, our assumption is that such variables are likely to be evenly (randomly) distributed or have minimal impact on the overall performance of the GMWI2 tool, given the breadth and reasonable diversity inherent in our study’s sample population. Last, our definitions of healthy (i.e., self-reported absence of a disease or disease-related symptoms) and non-healthy (i.e., patients with a clinical diagnosis of a disease) are consistent with those used in our previous studies 10 , 11 , as the current work represents a continuous refinement of our previous method. However, we have not investigated how subtle variations in these definitions may impact GMWI2 classification accuracy. Analyzing this aspect is a potential area for future research.
In regard to its translational potential, GMWI2 is designed to offer a novel method for dynamically monitoring an individual’s health in a semi-real-time manner through the analysis of gut microbiome taxonomic profiles. While our index is explicitly trained to distinguish between healthy and diseased gut microbiomes, it also provides a practical approach to approximating pre-diseased states. This is achieved by interpolating between the healthy and diseased states, allowing GMWI2 to reveal variations across the gut microbiome health spectrum. Specifically, assuming sufficient prediction quality of our model, an individual’s GMWI2 score will decrease as they transition from healthy to pre-diseased to diseased states, or increase if transitioning in the reverse direction. Moreover, GMWI2 provides a pragmatic alternative to the resource-intensive collection of longitudinal gut microbiome datasets needed to precisely track the steady transition from healthy to diseased. Current efforts in this area are very limited in scale and costly.
In all, GMWI2 is not intended for confirming specific disease diagnoses but rather serves as an early warning system, akin to a “canary in a coal mine”. It is designed to detect potentially adverse shifts in overall gut health before specific, diagnosable symptoms occur. Such detection could inform dietary or lifestyle modifications to prevent mild issues from escalating into severe health conditions, or prompt further diagnostic tests. Unlike existing disease-specific indices, our index spans multiple diseases, thereby emphasizing a pan-disease (or alternatively, a generally healthy) gut microbiome signature. This broad applicability could be particularly useful in clinical scenarios such as selecting FMT donors, where gut health could be taken as a reflection of overall health. In conditions like rheumatoid arthritis and other autoimmune inflammatory disorders, GMWI2 could guide decisions on tapering or discontinuing therapy, or assessing the possibility of disease flares. In this sense, GMWI2 may potentially usher in a transformative era in gut microbiome-centric health analytics, allowing for nuanced health evaluations tailored to individual microbial signatures. Looking ahead, integrating GMWI2 into a larger decision network alongside other biomeasurements (e.g., multi-omics, wearables) and AI models has the potential to open exciting possibilities for healthy aging 43 and preventative health screening and wellness programs 44 , 45 , driven by insights from the gut microbiome.
Multi-study pooling of human stool metagenomes
We conducted a comprehensive literature search using targeted keywords such as “gut microbiome”, “stool metagenome”, and “whole-genome shotgun” in PubMed and Google Scholar. The search was performed up until January 2022 to identify published studies that included publicly available shotgun metagenomic data of human stool samples, along with corresponding subject meta-data. In cases where multiple samples were collected from individuals across different time points, we included only the first or baseline sample from that study subject. Studies involving dietary or medication interventions were not included in the pooled dataset for GMWI2 training. Studies with fewer than 40 samples were also excluded from our analysis, considering the potential limitations in the robustness and reliability of microbiome data from such pilot-scale microbiome studies. The raw sequence files (in .sra or .fastq format) were retrieved from the NCBI Sequence Read Archive and European Nucleotide Archive databases for further analysis.
Stool metagenome sample exclusion criteria
To minimize potential bias and preserve data integrity, we applied stringent criteria to the stool shotgun metagenome samples for inclusion in our study. Specifically, we excluded samples sequenced using non-Illumina platforms, such as 454 GS FLX Titanium, Ion Torrent PGM, Ion Torrent Proton, and BGISEQ-500, to ensure consistency in sequencing technology. In terms of data quality, we excluded samples with low read counts (below 1 million reads) prior to quality control filtration. Additionally, our analysis did not include samples from studies with a primary focus on the virome or those where stool samples underwent virus-like particle purification.
Furthering our strict sample control standards, we also excluded disease control samples that were not specifically tied to a clinical diagnosis in the originating study. Individuals who were not clinically diagnosed with a specific disease but exhibited certain anomalous conditions were also excluded. These conditions comprised: (i) a Body Mass Index (BMI) suggestive of being underweight (BMI < 18.5), overweight (BMI ≥ 25 and <30), or obese (BMI ≥ 30) were not classified as a non-healthy phenotype; (ii) declared heavy drug use (including alcohol and recreational drugs); (iii) age exceeding 100 years; and (iv) individuals initially healthy at baseline, but later reported to develop a disease condition during a longitudinal study. Additionally, samples from newborn, infant, and child gut microbiome studies were excluded since the primary focus was on adult human gut microbiomes. Lastly, we excluded non-healthy individuals with early-stage diseases (e.g., impaired glucose tolerance, hypertension, colorectal adenoma), rare or genetically-linked disorders (e.g., Behcet’s disease, schizophrenia), and non-colon cancers (including pancreatic, non-small cell lung, and breast cancer). These exclusions were applied to ensure a uniform and representative dataset for training GMWI2.
Quality control of sequenced reads
Potential human contamination was filtered out by removing reads that aligned to the human genome (reference genome GRCh38/hg38) using Bowtie2 46 v2.4.4 with default parameters. Along with Illumina universal adapter sequences, probable adapter sequences were identified by extracting overrepresented sequences from each metagenome sample using FastQC 47 v0.11.8. Adapter sequence clipping and quality filtration were performed using Trimmomatic 48 v0.39. Specifically, Trimmomatic’s “ILLUMINACLIP” step was used, using a maximum seed mismatch count of 2, palindrome clip threshold of 30, simple clip threshold of 10, and minimum adapter length of 2 bp. Additionally, leading and trailing low-quality bases (Phred quality score < 3) of each read were removed, and trimmed reads shorter than 60 bp in nucleotide length were discarded.
Taxonomic profiling
After performing quality filtration on all raw metagenomes, taxonomic profiling was carried out using the MetaPhlAn3 16 v3.0.13 phylogenetic clade identification pipeline using default parameters. Briefly, MetaPhlAn3 classifies metagenomic reads to taxonomies based on a database (mpa_v30_CHOCOPhlAn_201901) of clade-specific marker genes. Once taxonomic features (or clades) of unknown/unclassified identity were removed, the remaining clades that could be detected in at least one metagenome sample in the pooled dataset were considered for further analysis.
After taxonomic profiling, the following metagenomes were discarded from our analysis: (i) samples composed of >90% unmapped reads; (ii) samples with a relatively high proportion (>25%) of unknown taxa; and (iii) samples lacking sufficient taxonomic diversity (<100 identified taxa). These samples were removed to maintain the quality and reliability of the training data. Finally, after applying all exclusion criteria, studies with fewer than 20 remaining samples were removed.
Generating presence/absence taxonomic profiles
To mitigate concerns related to the compositional nature of microbiome data 49 , batch effects, and to simplify the interpretation of the GMWI2 classification model, we transformed the taxa relative abundances from MetaPhlAn3 into a binary presence/absence profile for each metagenome sample. Specifically, a taxon was deemed “present” in a given sample if its relative abundance in a sample was equal to or greater than 0.00001 (or 0.001%), and considered absent otherwise. Consequently, each sample was represented as a binary vector.
PCA and PERMANOVA analysis on taxonomic profiles
Principal component analysis (PCA) was conducted on the presence/absence taxonomic profiles using the “prcomp” function in R. Additionally, Bray-Curtis distance matrices were generated based on the relative abundances of microbial taxa (ranging from phylum to species) in the stool metagenomes. This was done using the “vegan” package v2.6.4 in R. We then carried out permutational multivariate analysis of variance (PERMANOVA) on the distance matrix using the “adonis2” function. To evaluate the influence of the subjects’ health status on the total variance in gut microbial community composition, we calculated the P -value for the test statistic (pseudo-F) based on 999 permutations.
Estimating disease likelihood using Lasso-penalized logistic regression
A Lasso-penalized logistic regression model (Python library “scikit-learn” v1.0.2) was trained on the binary presence/absence taxonomic profiles of the entire pooled dataset of 8069 metagenomes to predict disease presence. The L1 (Lasso) penalty was utilized with the LIBLINEAR solver 50 . The random state was set to 42, and the class weight was set to “balanced” in order to account for the unbalanced class proportions in our pooled dataset. Hyperparameter tuning—specifically the selection of the regularization parameter \(C\) —was achieved through nested cross-validation that implements the inter-study validation (ISV) framework. Herein, we evaluated various candidates and selected the value that yielded the optimal classification performance in ISV (Supplementary Table 9 ; see table footnote for our nested cross-validation protocol). \(C\) = 0.03 consistently emerged as the optimal hyperparameter within each outer-loop training fold and was thus selected for the final GMWI2 model.
Let \({{{\boldsymbol{x}}}}_{i}\) be a binary vector encoding the presence or absence of n taxa in the i th labeled sample:
where \({x}_{i}^{j}\) is 1 if taxa \(j\) is present in sample \(i\) and 0 otherwise. Additionally, n = 3200 is the number of taxonomic features (or clades) in the i th sample (a total of 3200 taxonomic features were observed at least once in the pooled metagenome dataset).
Let \({y}_{i}\) represent the health status (1 for healthy, 0 for non-healthy) of sample i . The subsequent log-loss optimization objective function is solved using L1 regularization and class proportion weights as follows:
where \({\theta }^{*}\) refers to the learned coefficient vector, \(C\) is the aforementioned inverse regularization strength parameter, m = 8069 represents the total number of samples in the pooled metagenome dataset, \(\alpha\) is the class proportion weight term, and \({h}_{\theta }({{{\boldsymbol{x}}}}_{i})\) is the hypothesis function:
where \(\sigma\) is the sigmoid function. The class proportion term \(\alpha\) accounts for the relatively unbalanced class proportions in the pooled dataset:
Using GMWI2 as a stool metagenome-based health status classifier
We calculated GMWI2 scores for all 8069 stool metagenomes in the pooled dataset, as well as samples from the four gut microbiome case studies. The taxonomic profile of a metagenome was represented as a vector \({{{\boldsymbol{x}}}}_{{\mbox{test}}}\) , with binary values that encoded the presence or absence of microbial taxa. The computation employed the predicted log odds (logit) using the previously learned coefficient vector \({\theta }^{*}\) :
For classification purposes, a predetermined magnitude cutoff parameter \(c\) was utilized ( \(c=\,0\) in case of having no cutoff or defer option). Finally, GMWI2 was computed on a metagenome \({{{\boldsymbol{x}}}}_{{test}}\) while applying the following criteria:
Of note, our current methodology does not inherently categorize gut microbiome samples into a third option. GMWI2 yields a continuous score, where the sign (negative or positive) is indicative of disease presence or absence, respectively; and higher magnitudes imply greater confidence in the prediction. The “defer” (or “not determined”) category is an optional feature, applicable when a user decides to implement a non-zero GMWI2 magnitude cutoff \(c\) . Scores falling below this user-defined cutoff (e.g., between –1.0 and +1.0) can be classified as “defer.”
Evaluation of classification performance
Balanced accuracy, defined as the average of the proportions of correctly classified healthy and non-healthy samples, was used to evaluate the performance of the GMWI2 classification model. This was done across different cutoff parameters ( c ) using multiple validation techniques: training on the entire dataset and then testing on the same training set, 10-fold cross-validation (10-fold CV), and leave-one-out cross-validation (LOOCV). In order to account for variability in 10-fold cross-validation, the process was repeated 10 times with shuffled fold partitions, and the results were averaged across all runs. Additionally, inter-study validation (ISV) was conducted, in which a single study was held out each time, the model was trained on the remaining studies, and testing was performed on the samples of the single-held-out study. ISV allows for an assessment of classification performance across different studies.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Information regarding the stool metagenome samples (and their corresponding studies) used to train the GMWI2 classifier is available in Supplementary Data 2 . Raw metagenomic reads are available using the sequencing data accession IDs.
Code availability
A command-line tool for computing the GMWI2 score of a stool metagenome from its corresponding raw .fastq sequence file can be installed via Anaconda ( https://anaconda.org/bioconda/GMWI2 ). The source code for the tool, processed datasets (including the taxonomic profiles of all metagenome samples analyzed in this study), and code notebooks essential to reproduce all results presented in our study, as well as complete instructions for installation and usage, are freely available online at https://github.com/danielchang2002/GMWI2 .
Schirmer, M. et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 167 , 1125–1136.e8 (2016).
Article CAS PubMed PubMed Central Google Scholar
Halfvarson, J. et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2 , 1–7 (2017).
Article Google Scholar
Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569 , 655–662 (2019).
Article ADS CAS PubMed PubMed Central Google Scholar
Wirbel, J. et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 25 , 679–689 (2019).
Mars, R. A. T. et al. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell 183 , 1137–1140 (2020).
Article CAS PubMed Google Scholar
Mou, Y. et al. Gut Microbiota interact with the brain through systemic chronic inflammation: Implications on neuroinflammation, neurodegeneration, and aging. Front. Immunol. 13 , 796288 (2022).
iMSMS Consortium. Gut microbiome of multiple sclerosis patients and paired household healthy controls reveal associations with disease risk and course. Cell 185 , 3467–3486.e16 (2022).
Ferreiro, A. L. et al. Gut microbiome composition may be an indicator of preclinical Alzheimer’s disease. Sci. Transl. Med. 15 , eabo2984 (2023).
Morton, J. T. et al. Multi-level analysis of the gut-brain axis shows autism spectrum disorder-associated molecular and microbial profiles. Nat. Neurosci. 26 , 1208–1217 (2023).
Gupta, V. K. et al. A predictive index for health status using species-level gut microbiome profiling. Nat. Commun. 11 , 4635 (2020).
Chang, D., Gupta, V. K., Hur, B., Cunningham, K. Y. & Sung, J. GMWI-webtool: a user-friendly browser application for assessing health through metagenomic gut microbiome profiling. Bioinformatics 39 , btad061 (2023).
Gacesa, R. et al. Environmental factors shaping the gut microbiome in a Dutch population. Nature 604 , 732–739 (2022).
Article ADS CAS PubMed Google Scholar
Xu, Q. et al. Metagenomic and metabolomic remodeling in nonagenarians and centenarians and its association with genetic and socioeconomic factors. Nat. Aging 2 , 438–452 (2022).
Knights, D., Parfrey, L. W., Zaneveld, J., Lozupone, C. & Knight, R. Human-associated microbial signatures: examining their predictive value. Cell Host Microbe 10 , 292–296 (2011).
Pasolli, E., Truong, D. T., Malik, F., Waldron, L. & Segata, N. Machine learning meta-analysis of large metagenomic datasets: tools and biological insights. PLoS Comput. Biol. 12 , e1004977 (2016).
Article ADS PubMed PubMed Central Google Scholar
Beghini, F. et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife 10 , e65088 (2021).
Truong, D. T. et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12 , 902–903 (2015).
Leek, J. T. et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 11 , 733–739 (2010).
Sung, J., Wang, Y., Chandrasekaran, S., Witten, D. M. & Price, N. D. Molecular signatures from omics data: from chaos to consensus. Biotechnol. J. 7 , 946–957 (2012).
Pattee, J. & Pan, W. Penalized regression and model selection methods for polygenic scores on summary statistics. PLoS Comput. Biol. 16 , e1008271 (2020).
Choi, S. W., Mak, T. S.-H. & O’Reilly, P. F. Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc. 15 , 2759–2772 (2020).
Ma, S. et al. Measuring the effect of inter-study variability on estimating prediction error. PLoS ONE 9 , e110840 (2014).
Goll, R. et al. Effects of fecal microbiota transplantation in subjects with irritable bowel syndrome are mirrored by changes in gut microbiome. Gut Microbes 12 , 1794263 (2020).
Article PubMed PubMed Central Google Scholar
Woodworth, M. H., Carpentieri, C., Sitchenko, K. L. & Kraft, C. S. Challenges in fecal donor selection and screening for fecal microbiota transplantation: a review. Gut Microbes 8 , 225–237 (2017).
Duvallet, C. et al. Framework for rational donor selection in fecal microbiota transplant clinical trials. PLoS ONE 14 , e0222881 (2019).
Tanes, C. et al. Role of dietary fiber in the recovery of the human gut microbiome and its metabolome. Cell Host Microbe 29 , 394–407.e5 (2021).
Gibson, G. R. & Roberfroid, M. B. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125 , 1401–1412 (1995).
Anderson, J. W. et al. Health benefits of dietary fiber. Nutr. Rev. 67 , 188–205 (2009).
Article PubMed Google Scholar
Venter, C. et al. Role of dietary fiber in promoting immune health—An EAACI position paper. Allergy 77 , 3185–3198 (2022).
Palleja, A. et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 3 , 1255–1265 (2018).
Lee, D. H. et al. Evaluating the prebiotic effect of oligosaccharides on gut microbiome wellness using in vitro fecal fermentation. Npj Sci. Food 7 , 18 (2023).
Zeevi, D. et al. Personalized nutrition by prediction of glycemic responses. Cell 163 , 1079–1094 (2015).
Ananthakrishnan, A. N. et al. Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe 21 , 603–610.e3 (2017).
Hjorth, M. F. et al. Prevotella-to-Bacteroides ratio predicts body weight and fat loss success on 24-week diets varying in macronutrient composition and dietary fiber: results from a post-hoc analysis. Int. J. Obes. 43 , 149–157 (2019).
Article CAS Google Scholar
Gupta, V. K. et al. Gut microbial determinants of clinically important improvement in patients with rheumatoid arthritis. Genome Med. 13 , 149 (2021).
Wilmanski, T. et al. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat. Metab. 3 , 274–286 (2021).
Jian, C. et al. Gut microbiota predicts body fat change following a low-energy diet: a PREVIEW intervention study. Genome Med . 14 , 54 (2022).
Sung, J. et al. Multi-study integration of brain cancer transcriptomes reveals organ-level molecular signatures. PLoS Comput. Biol. 9 , e1003148 (2013).
Parsana, P., Amend, S. R., Hernandez, J., Pienta, K. J. & Battle, A. Identifying global expression patterns and key regulators in epithelial to mesenchymal transition through multi-study integration. BMC Cancer 17 , 447 (2017).
Xu, J. et al. Algorithmic fairness in computational medicine. EBioMedicine 84 , 104250 (2022).
Herbei, R. & Wegkamp, M. H. Classification with Reject Option. Can. J. Stat. 34 , 709–721 (2006).
Article MathSciNet Google Scholar
Hanczar, B. & Dougherty, E. R. Classification with reject option in gene expression data. Bioinformatics 24 , 1889–1895 (2008).
Ghosh, T. S., Shanahan, F. & O’Toole, P. W. Toward an improved definition of a healthy microbiome for healthy aging. Nat. Aging 2 , 1054–1069 (2022).
Price, N. D. et al. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat. Biotechnol. 35 , 747–756 (2017).
Shen, X. et al. Multi-omics microsampling for the profiling of lifestyle-associated changes in health. Nat. Biomed. Eng . 8 , 1–19 (2024).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 , 357–359 (2012).
Wingett, S. W. & Andrews, S. FastQ Screen: a tool for multi-genome mapping and quality control. F1000Res. 7 , 1338 (2018).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 , 2114–2120 (2014).
Gloor, G. B., Macklaim, J. M., Pawlowsky-Glahn, V. & Egozcue, J. J. Microbiome datasets are compositional: and this is not optional. Front. Microbiol . 8 , 2224 (2017).
Fan, R.-E., Chang, K.-W., Hsieh, C.-J., Wang, X.-R. & Lin, C.-J. LIBLINEAR: a library for large linear classification. https://www.csie.ntu.edu.tw/~cjlin/papers/liblinear.pdf (2008).
Download references
Acknowledgements
We thank Professor Dan Knights for helpful feedback regarding this study. This work was supported in part by the National Center for Advancing Translational Sciences of the National Institutes of Health Award Numbers UL1TR002494 and UL1TR002377. Additional support was provided by the Minnesota Partnership for Biotechnology and Medical Genomics through the Translational Product Development Fund (to J.S.), as well as Mark E. and Mary A. Davis to Mayo Clinic Center for Individualized Medicine (to J.S.).
Author information
These authors contributed equally: Daniel Chang, Vinod K. Gupta.
Authors and Affiliations
Department of Computer Science and Engineering, University of Minnesota, Minneapolis, MN, USA
Daniel Chang
Microbiomics Program, Center for Individualized Medicine, Mayo Clinic, Rochester, MN, USA
Vinod K. Gupta, Benjamin Hur & Jaeyun Sung
Viral Information Institute, San Diego State University, San Diego, CA, USA
Sergio Cobo-López
Bioinformatics and Computational Biology Program, University of Minnesota, Minneapolis, MN, USA
Kevin Y. Cunningham
Brain Korea 21 Center for Bio-Health Industry, Department of Food Science and Biotechnology, Chungbuk National University, Cheongju, South Korea
Nam Soo Han
Department of Biotechnology, Yonsei University, Seoul, South Korea
Division of Rheumatology, Department of Medicine, Mayo Clinic, Rochester, MN, USA
Vanessa L. Kronzer, John M. Davis III & Jaeyun Sung
Department of Food Science and Nutrition, University of Minnesota, St. Paul, MN, USA
Levi M. Teigen
Department of Pulmonary & Critical Care, Mayo Clinic, Rochester, MN, USA
Lioudmila V. Karnatovskaia
Department of Neurology, Yale University, New Haven, CT, USA
Erin E. Longbrake
Emeritus, Department of Surgery, Mayo Clinic, Rochester, MN, USA
Heidi Nelson
Division of Computational Biology, Department of Quantitative Health Sciences, Mayo Clinic, Rochester, MN, USA
Jaeyun Sung
You can also search for this author in PubMed Google Scholar
Contributions
D.C., V.K.G. and J.S. developed the study idea and designed all analytical methodologies. D.C. and V.K.G. performed the computational experiments. All authors (D.C., V.K.G., B.H., S.C.-L., K.Y.C., N.H., I.L., V.L.K., L.M.T., L.V.K., E.E.L., J.M.D., H.N. and J.S.) analyzed and discussed the data. D.C., V.K.G. and J.S. wrote the manuscript, with contributions from other authors. All authors critically reviewed and approved the final manuscript.
Corresponding author
Correspondence to Jaeyun Sung .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Peer review
Peer review information.
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information, peer review file, description of additional supplementary files, supplementary data 1, supplementary data 2, supplementary data 3, supplementary data 4, supplementary data 5, supplementary data 6, supplementary data 7, reporting summary, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ .
Reprints and permissions
About this article
Cite this article.
Chang, D., Gupta, V.K., Hur, B. et al. Gut Microbiome Wellness Index 2 enhances health status prediction from gut microbiome taxonomic profiles. Nat Commun 15 , 7447 (2024). https://doi.org/10.1038/s41467-024-51651-9
Download citation
Received : 25 September 2023
Accepted : 09 August 2024
Published : 28 August 2024
DOI : https://doi.org/10.1038/s41467-024-51651-9
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Microbiology newsletter — what matters in microbiology research, free to your inbox weekly.
Official websites use .gov
A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS
A lock ( ) or https:// means you've safely connected to the .gov website. Share sensitive information only on official, secure websites.
SNAP Emergency Allotments, Emergency Rent Assistance, Rent Burden, and Housing and Food Security, June 2022–May 2023
ORIGINAL RESEARCH — Volume 21 — August 29, 2024
Patrick J. Brady, PhD 1 ; Kaitlyn M. Berry, PhD, MPH 1 ; Rachel Widome, PhD, MHS 1 ; Sruthi Valluri, MD, PhD 2 ; Melissa N. Laska, PhD, RD 1 ( View author affiliations )
Suggested citation for this article: Brady PJ, Berry KM, Widome R, Valluri S, Laska MN. SNAP Emergency Allotments, Emergency Rent Assistance, Rent Burden, and Housing and Food Security, June 2022–May 2023. Prev Chronic Dis 2024;21:240121. DOI: http://dx.doi.org/10.5888/pcd21.240121 .
PEER REVIEWED
Introduction
Acknowledgments, author information.
What is already known on this topic?
Economic supports implemented in the US during the COVID-19 pandemic improved material conditions in US households, but we do not know if improvements differed according to a household’s economic situation.
What is added by this report?
The association between receiving Supplemental Nutrition Assistance Program emergency allotments or emergency rent assistance and indicators of food or housing security differed according to whether households were experiencing rent burden, sometimes with stronger, positive associations for non–rent-burdened households.
What are the implications for public health practice?
Policies that support public health and prevent chronic disease by addressing social determinants of health should be purposefully designed to equitably address the needs of economically constrained households.
During the COVID-19 pandemic, Supplemental Nutrition Assistance Program (SNAP) emergency allotments and emergency rent assistance provided support to low-income households. Rent burden, a form of housing insecurity, can severely limit household resources, which, in turn, affects health equity. We explored whether these policy interventions equitably supported households that were or were not experiencing rent burden.
We used data from the US Household Pulse Survey (June 2022–May 2023) to examine whether associations between emergency support policies and indicators of food and housing security differed according to household rent burden status. We modeled each outcome (food sufficiency or being current on rent) as a function of policy exposure (SNAP emergency allotments or emergency rent assistance), rent burden, and their interaction. We included demographic characteristics, state of residence, and survey cycle as covariates. We modeled each outcome and policy exposure combination separately.
Receiving emergency allotments (72.4% vs 67.2% for SNAP participants in states with and without emergency allotments, respectively) and emergency rent assistance (64.5% vs 57.6% for households that received and were waitlisted/denied assistance, respectively) were associated with greater food sufficiency. The relationship between emergency allotments and food sufficiency was stronger in rent-burdened households; however, emergency rent assistance supported food sufficiency to a greater extent in non–rent-burdened households. Emergency rent assistance supported households in being current on rent (78.7% vs 56.4% for households that received and were waitlisted/denied assistance, respectively) and supported being current on rent to a greater extent in non–rent-burdened households than in rent-burdened households.
The relationship between emergency support policies and food or housing security differed according to whether households were experiencing rent burden. Associations were sometimes stronger in less economically constrained conditions. These results indicate an opportunity to better design policies to support low-income households, address food and housing security, and ultimately decrease the prevalence of chronic disease.
The COVID-19 pandemic exposed the inadequacy of the US social safety net. However, a robust policy response supported households through many mechanisms, including direct economic support (eg, enhanced child tax credit, economic impact payments, enhanced unemployment insurance), a wider food safety net (increases in Supplemental Nutrition Assistance Program [SNAP] benefits, SNAP emergency allotments, and Pandemic EBT [electronic benefits transfer] to replace school meals), and an enhanced housing safety net (emergency rental assistance, eviction moratoriums) (1–8). A growing body of research on the policy response to the pandemic provides evidence that state provision of economic support to low-income households improves their material security (2–8). Policies that meet economic needs are therefore a major part of the solution to the problems of material insecurities that drive health inequities in preventable chronic diseases.
Food security, or “access by all people at all times to enough food for an active, healthy life” (9), is recognized as a critical social determinant of health, and lack of access to adequate food is a key contributor to the prevalence of and disparities in chronic disease (10,11). Inadequate food access drives risk of and disparities in nutrition-related chronic disease outcomes, such as for cardiovascular diseases and type 2 diabetes (10,11). Substantial evidence indicates that SNAP, which provides in-kind support to purchase eligible foods from authorized retailers, supports food security (12). SNAP emergency allotments were enacted in 2020 as a temporary measure that provided the maximum benefit amount for SNAP participants during the COVID-19 public health emergency (13). Before April 2021, households already receiving the maximum allowable amount, the most economically constrained participants, were excluded from receiving additional support (13). Beginning in April 2021, SNAP emergency allotments provided at least an additional $95 per household per month for households receiving the maximum allowable amount (3,4,13). This revised policy ensured that all SNAP participants in states implementing emergency allotments received additional benefits (13). Eighteen states chose to end SNAP emergency allotments early before the program expired in February 2023. Ending emergency allotments reduced benefits by an estimated average of $90 per person per month (13) and increased the number of households experiencing food insufficiency in the previous week from 25% to 31% (3).
Another key social determinant of health, housing security, is the “availability of and access to stable, safe, adequate, and affordable housing and neighborhoods regardless of gender, race, ethnicity, or sexual orientation” (14). During the COVID-19 pandemic, approximately 16% of households were behind on their rent, and this disproportionality affected racially and ethnically minoritized households (15). A critical dimension of housing security is cost burden, or the amount of income put toward housing costs (14,16,17). Renters who spend more than 30% of their income on rent are considered rent burdened (17,18). Rent burden is increasing in the US and is now recognized as a pressing public health issue (16,19). Housing unaffordability and rent burden are associated with negative social and economic outcomes (20), including food insecurity (21–24). Housing insecurity drives chronic disease outcomes and disparities such as food insecurity (11) by determining the food and health environment in which a household exists and by contributing to resource constraint. Addressing housing insecurity may support food security (25–27). One intervention to reduce housing insecurity is emergency rent assistance, which often directly covers unpaid rent owed to the landlord and/or unpaid utility costs. Many new state and local emergency rent assistance programs emerged early in 2020 and throughout the pandemic (7). These programs were often funded through federal legislation (28,29) and represent the first widescale implementation of this kind of housing assistance. Emergency rent assistance has been shown to reduce eviction filings (5), decrease the financial burden associated with housing insecurity (6,8), and improve mental health (6,8). Despite the beneficial effects of emergency rent assistance (5–8), these programs encountered implementation challenges and were not designed to address long-standing, systemic issues around housing affordability (30–32).
Generally, we know that SNAP emergency allotments (3,4) and emergency rent assistance (5–8) had positive effects. These effects, however, may have differed according to the material context of households. The objective of our study was to examine whether the association between receiving SNAP emergency allotments or emergency rent assistance and indicators of food and housing security differed according to whether households were experiencing rent burden. Based on previous literature (3,4,6,8,18), we hypothesized that experiencing rent burden would decrease food and housing security among low-income renters, whereas receiving SNAP emergency allotments or receiving emergency rent assistance would increase food and housing security. We also hypothesized that a significant interaction would exist between experiencing rent burden and each policy exposure. We did not hypothesize the direction of the interaction effect, evaluating it only in an exploratory manner.
We used data from the nationally representative US Census Bureau Household Pulse Survey (HPS) from June 2022 through May 2023, which included cycles 46 to 57. We selected these cycles because of the availability of the variable (monthly rental cost) needed to evaluate experiences of rent burden. More details on the HPS methodology are published elsewhere (33). Briefly, the HPS is a repeated cross-sectional survey designed to assess household social and economic conditions. Households are sampled from the US Census Bureau’s Master Address File, and information from the US Census Bureau’s Contact Frame enables contact with respondents through email and text messages. A single respondent answers questions about their household. Survey cycles typically lasted about 2 weeks. For the cycles used in our analysis, a gap of approximately 2 weeks occurred between each cycle. Survey weights account for survey design and nonresponse to generate demographically representative national-level estimates based on educational attainment, sex, age, race, and ethnicity.
Our sample consisted of renters with incomes less than 130% of the federal poverty threshold (N = 40,895). We used the self-reported categorical measure of yearly income and household size (number of household members) to determine eligibility based on income. If a household in any income category was under 130% of the federal poverty threshold, we included that household in the under-130% category. For example, the federal poverty threshold for a 3-member household is $29,939. Thus, a 3-member household with a yearly income of either less than $25,000 or $25,000 to $34,999 would be included in the under-130% category. Likewise, households with a yearly income of $50,000 to $74,999 would need to include at least 7 members to be considered a low-income renter for the purposes of our study.
Our outcomes of interest were food sufficiency and being current on rent. Food sufficiency, a dimension of food security, describes households with enough food for all household members (10). We assessed food sufficiency with a single item that asked households about the amount and types of foods they had available to eat in the past week. Households were considered “food sufficient” if they had enough of the kind of foods or enough but not always the kinds of food they wanted to eat; “food insufficient” described households that sometimes or often did not have enough to eat. Being unable to make rent payments is a housing affordability dimension of housing insecurity and is a more severe measure of housing affordability than experiencing rent burden (14). Being current on rent payments was assessed with a single item that asked, “Is this household currently caught up on rent payments?” Response options were yes or no.
We were interested in exposure to 2 policies: SNAP emergency allotments and emergency rent assistance. Because some states discontinued SNAP emergency allotments before the program expired in February 2023, households that were exposed to this policy can be identified according to participation, state of residence, and time period (3,4). We used indicator variables for exposure to emergency allotments based on self-reported SNAP participation at the time of the survey, state of residence, and survey cycle in a similar manner to previously reported analyses (3,4). This process resulted in a 4-level variable: 1) SNAP participant in a state with emergency allotments, 2) SNAP participant in a state without emergency allotments, 3) non–SNAP participant in a state with emergency allotments, and 4) non–SNAP participant in a state without emergency allotments. Our indicator for emergency rent assistance was based on the self-reported answer to the question “Have you or anyone in your household applied for emergency rental assistance through your state or local government to cover your unpaid rent or utility bills?” Response options were “Yes, received emergency rent assistance,” “No, waitlisted or denied emergency rent assistance,” or “No, did not apply.” Our moderator was experiencing rent burden (yes or no). We coded a household as experiencing rent burden if yearly rent costs (calculated from current monthly rent at the time of the survey) divided by the midpoint of income category was 0.30 or greater.
We controlled for demographic characteristics known to be associated with socioeconomic status. These were age category of respondent (≤35, 36–64, or ≥65 y), presence of children in household (yes or no), gender identity of respondent (male, female, transgender or other than any listed option), racial self-classification of respondent (Asian, Black, White, or any other race alone or any combination of responses), self-classification of respondent as having Hispanic, Latino, or Spanish origin (yes or no), yearly household income (<$25,000, $25,000–$34,999, $35,000–$49,999, or $50,000–$74,999), if anyone in household experienced a job loss in the past 4 weeks (yes or no), if anyone in household had employment in the past week (yes or no), educational attainment of respondent (less than high school or some high school, high school graduate or equivalent, some college but degree not received or in progress, associate or bachelor’s degree, graduate degree), marital status (now married, widowed, divorced, separated, never married), and if a household received food aid. The yes–no question about food aid asked if, during the last 7 days, anyone in the household got free groceries from a food pantry, food bank, church, or other place that helps with free food. Gender identity, racial self-classification, and Hispanic self-classification variables were included as proxies for experiences of structural and systemic discrimination that affect food, housing, and economic security. Finally, we included fixed effects for state (each of the 50 states and the District of Columbia) and the survey cycle.
First, we produced descriptive statistics for all variables overall and stratified by policy exposure. We also descriptively examined our outcomes during the study period. Then, we estimated the main effects of each policy exposure and the moderator on our outcomes of interest. Specifically, we used logistic regressions to model the outcomes (household food sufficiency or being current on rent) as a function of receiving SNAP emergency allotments, receiving emergency rent assistance, or experiencing rent burden. We adjusted models for demographic characteristics, state, and survey cycle. We tested separately the effect of each exposure or moderator on each outcome, resulting in 6 main effect regression models. From each model, we calculated the predicted probability of experiencing the outcome for each level of the exposure–moderator variable. We then calculated the difference in the predicted probability of the outcome across exposure–moderator levels.
We then ran interaction models to test whether the effect of each policy exposure differed according to whether households were rent burdened. Specifically, we estimated the outcomes (food sufficiency or being current on rent) as a function of receiving SNAP emergency allotments or emergency rent assistance, experiencing rent burden, and the interaction between policy exposure and rent burden. Each exposure–moderator interaction was tested separately. As with the main effects models, we then calculated the predicted probability of experiencing the outcome for each level of the interaction. We then calculated the differences in predicted probabilities for key comparisons of interest, using contrast statements following the interaction model.
Finally, we wanted to examine the effect of the decision to use the midpoint of each income category when determining whether a household experienced rent burden. Therefore, we conducted a sensitivity analysis where a household was designated rent burdened if their yearly rent costs divided by the top level of their income category was 0.30 or greater (ie, assuming each household had the highest possible income for their income category instead of the midpoint). We used Stata version 17 (StataCorp LLC) for all analyses. All analyses were weighted as described in HPS technical documentation (33).
In the overall sample, 83.4% reported being rent burdened, 69.2% food sufficient, and 80.4% current on rent ( Table 1 ). Overall, 63.7% were White, 23.8% were Black, and 24.2% were of Hispanic, Latino, or Spanish origin; 62.4% of respondents identified as female. The annual household income category with the largest percentage (70.6% of respondents) was less than $25,000; 41.6% had at least a high school diploma, and 17.7% received food aid. The mean (SD) household size was 3.0 (1.5) members. The rates of households experiencing food sufficiency and being current on rent were relatively consistent during the study period, ranging from 65.6% to 72.4% (food sufficiency) and 76.8% to 83.0% (current on rent) ( Figure ).
Food sufficiency
In the main effects models, food sufficiency was less common among households that experienced rent burden than among those that did not (68.7% vs 71.7%), higher among SNAP participants who received emergency allotments than among those that did not (72.4% vs 67.2%), and higher among households that received emergency rent assistance than among those who were waitlisted or denied (64.5% vs 57.6%) ( Table 2 ). In the interaction model for SNAP emergency allotments and rent burden, emergency allotments supported food sufficiency when SNAP participating households were rent burdened (72.0% vs 66.0%) but not if they were non–rent burdened (74.0% vs 71.5%) ( Table 2 ). In the interaction model for emergency rent assistance and rent burden, receiving assistance supported food sufficiency to a greater extent when households who applied for assistance were non–rent burdened (68.9% vs 55.9% for non–rent-burdened households and 63.0% vs 57.8% for rent-burdened households) ( Table 2 ).
Current on rent
In the main effects models, being current on rent was less common among households that experienced rent burden than among non–rent-burdened households (79.5% vs 85.1%) ( Table 3 ). The predicted probability of being current on rent was similar across SNAP participants and non–SNAP participants regardless of policy exposure. Among households that applied for emergency rent assistance, those who received assistance (78.7%) were substantially more likely to be current on rent than those who were waitlisted or denied (56.4%) ( Table 3 ). In the interaction model for SNAP emergency allotments and rent burden, we found no clear relationship between experiencing rent burden and being exposed to SNAP emergency allotments. In the interaction model for emergency rent assistance and rent burden, assistance supported being current on rent to a greater extent when households were non–rent burdened (85.5% vs 60.9% for non–rent-burdened households and 76.7% vs 55.8% for rent-burdened households) ( Table 3 ).
Sensitivity analysis
In the sensitivity analysis, assuming that each household had the highest possible income for their income category, instead of the midpoint, the main effect of rent burden was diminished, but trends in the interaction models were similar.
We found that while both SNAP emergency allotments and emergency rent assistance supported household economic security, the association between receiving these benefits and food or housing security was affected by whether the household was rent burdened. Receiving SNAP emergency allotments was more strongly associated with food sufficiency in rent-burdened households than in non–rent-burdened households. In contrast, emergency rent assistance was more strongly associated with food sufficiency and being current on rent in non–rent-burdened households than in rent-burdened households. Together, these findings suggest 2 possible mechanisms for the interaction between economic supports and experiencing rent burden. Programs that provide in-kind support may have a stronger effect in more economically constrained households because the resources will have a larger relative contribution to their financial capabilities. At the same time, households with fewer material constraints may be able to use additional resources more effectively to meet their needs. Future research, especially mixed-methods studies that can quantitatively assess resource allocation and qualitatively characterize participant experiences and perceptions driving those decisions, should examine these mechanisms.
Consistent with previous research (3,4), in our analysis SNAP emergency allotments supported the food sufficiency of households. We extended this research by documenting the differential effects of household rent burden. That emergency allotments had more effect among rent-burdened households than among non–rent-burdened households provides further support that increasing SNAP benefits could be a key component in supporting food security. These results also support a need to examine how policies interact with social, political, and economic marginalization to produce inequities and potentially drive health disparities. Future research should examine how the effects described here vary by demographic characteristics (eg, racial and ethnic identity, gender identity, household composition) and environmental contexts (eg, housing markets, food environments); whether the observed relationships are consistent across other policies that provide economic support; and how these contribute to a disparate burden of chronic disease on economically marginalized populations.
Although our analysis adds to the evidence on the effects of SNAP emergency allotments, these allotments were designed as a temporary measure. Providing the full benefit amount to all SNAP-participating households may not be a feasible long-term solution, and other mechanisms that increase benefits should be explored. A policy lever that may be of particular interest for simultaneously addressing food and housing security is the SNAP excess shelter deduction (34), which allows households applying for SNAP to claim a federally capped portion of their housing-related costs when determining net income. This deduction affects both eligibility and benefits: eligibility for SNAP is determined by net income test (at or below 100% of the federal poverty threshold) and net income is used in benefits computation (34). The excess shelter deduction could be more actively promoted to increase awareness or improved by removing administrative burdens to apply or increasing associated benefits. Some efforts to improve the excess shelter deduction have been proposed; the Closing the Meal Gap Act (35) would eliminate the deduction’s cap.
Our study results also have important implications for building on the success of emergency rental assistance programs implemented during the COVID-19 pandemic. Our results show that pandemic-related emergency rent assistance programs supported the housing security of non–rent-burdened households to a greater extent than it supported rent-burdened households. This finding could be due to assistance being more effective at addressing economic outcomes among non–rent-burdened households, which are likely less resource-constrained than rent-burdened households. Relatedly, rent-burdened households may be allocating any freed-up resources to more pressing needs, such as transportation, health care and prescription drugs, or childcare, rather than housing. Further research examining how households receiving emergency rent assistance allocate their resources before and after receiving benefits could provide more insight into these mechanisms. Such research could then be used to enhance rental assistance program design (eg, reaching those who are in most need or linking with other forms of support).
We also found that while emergency rent assistance supported both food and housing security, SNAP emergency allotments supported only food security, perhaps because of the relatively meager benefits offered by SNAP overall and, by extension, emergency allotments. Previous research showed that housing cost assistance can improve food security (25–27), but more research is needed to examine the effect, if any, of food assistance on housing security.
Overall, about 3 in 10 households in our analysis did not have enough food sometimes or often in the past week and nearly 20% were not current on rent payments. Additionally, even among non–rent-burdened households, a substantial number of households were facing housing insecurity and inadequate food access, highlighting the need for a more robust social safety net. Without addressing these social determinants of health, it will be impossible to reduce the incidence of preventable chronic diseases and reduce or eliminate disparities between populations. In a country with adequate resources to feed and house all, it is a societal failing that so many struggle with meeting their basic needs. Improving the social safety net, including supporting and advocating for a robust welfare state, should be a priority and emphasis should be placed on including those most affected by socially and politically manufactured food and housing scarcity (36).
Limitations
Our study has several potential limitations. We used specific dependent variables to reflect wide, multidimensional states of food and housing security. Additionally, our moderator was a measure of rent burden based on the percentage of income needed to pay rent, which does not fully capture the economic reality facing households. While other measures of rent burden, such as the residual income approach (17), more accurately reflect household resources, these measures require more detailed information on income and costs and therefore were not feasible for our analysis. Furthermore, because income data were reported categorically, we were unable to precisely identify households with incomes under 130% of the federal poverty threshold, and we calculated our moderator by using the midpoint of income categories, which may have misclassified some households. Additionally, we relied on self-report to establish receipt of benefits, which likely misclassified some households, with underreporting of receipt more likely than overreporting. There is also substantial selection into assistance programs based on a plethora of observable and unobservable factors, as demonstrated extensively in SNAP (12), for which our analysis did not account. This factor should be considered when comparing the relationship between SNAP participants and non-SNAP participants and households that applied for emergency rental assistance versus those that did not. Finally, our analysis focused on the moderating effect of rent burden and policies that economically supported households on indicators of food and housing security, but more advanced methods, such as difference-in-difference models, could be used to evaluate these relationships more fully.
The association between receiving economic support and outcomes related to food and housing security differs according to the rent-burdened status of households. This association did not always occur consistently: the association was stronger among rent-burdened households (vs non–rent-burdened households) for SNAP emergency allotments but stronger among non–rent-burdened households (vs rent-burdened households) for emergency rent assistance. Policies and programs that aim to address insecurity, such as food and housing assistance programs, should be designed to provide additional resources to households with fewer resources if those programs are shown to be more effective among less economically constrained households. Such designs could better address the social and economic conditions faced by resource-limited households and contribute to lessening the effect of economic inequities on chronic disease burden and disparities.
The authors declare no potential conflicts of interest with respect to the research, authorship, or publication of this article. P.J.B. is funded under award no. 1K99AG082884-01 from the National Institute on Aging (NIA). K.M.B. is funded under award no. F31HD107980 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and received resources and support from the Minnesota Population Center (P2CHD041023) also from NICHD. The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIA, NICHD, or National Institutes of Health. No copyrighted material, surveys, instruments, or tools were used in the research described in this article.
Corresponding Author: Patrick J. Brady, PhD, University of Minnesota School of Public Health, Ste 300, West Bank Office Bldg, 1300 South 2nd St, Minneapolis, MN 55455 ( [email protected] ).
Author Affiliations: 1 Division of Epidemiology and Community Health, University of Minnesota School of Public Health, Minneapolis, Minnesota. 2 Brigham and Women’s Hospital, Boston, Massachusetts.
- Béland D, Dinan S, Rocco P, Waddan A. Social policy responses to COVID-19 in Canada and the United States: explaining policy variations between two liberal welfare state regimes. Soc Policy Adm . 2021;55(2):280–294. PubMed doi:10.1111/spol.12656
- Shafer PR, Gutiérrez KM, Ettinger de Cuba S, Bovell-Ammon A, Raifman J. Association of the implementation of child tax credit advance payments with food insufficiency in US households. JAMA Netw Open . 2022;5(1):e2143296. PubMed doi:10.1001/jamanetworkopen.2021.43296
- Richterman A, Roberto CA, Thirumurthy H. Associations between ending Supplemental Nutrition Assistance Program emergency allotments and food insufficiency. JAMA Health Forum . 2023;4(8):e232511. PubMed doi:10.1001/jamahealthforum.2023.2511
- Sanjeevi N, Monsivais P. Association of emergency allotment discontinuation with household food insufficiency in Supplemental Nutrition Assistance Program participants: a quasi-experimental study. Prev Med . 2023;177:107784. PubMed doi:10.1016/j.ypmed.2023.107784
- Martin H. Making sense of eviction trends during the pandemic. Federal Reserve Bank of Cleveland. August 23, 2022. Accessed January 16, 2024. https://www.clevelandfed.org/publications/economic-commentary/2022/ec-202212-making-sense-of-eviction-trends-during-the-pandemic
- Reina VJ, Lee Y. COVID-19 and emergency rental assistance: impact on rent arrears, debt, and the well-being of renters in Philadelphia. RSF . 2023;9(3):208–229.
- Boshart A, Champion E, Popkin SJ. Pandemic rental assistance funding strengthened the renter safety net. Urban Institute, Housing Crisis Research Collaborative. February 2022. Accessed May 13, 2024. https://www.urban.org/sites/default/files/2022-03/pandemic-rental-assistance-funding-strengthened-the-renter-safety-net.pdf
- Airgood-Obrycki W. The Short-Term Benefits of Emergency Rental Assistance . Joint Center for Housing Studies of Harvard University. June 2022. Accessed May 13, 2024. https://www.jchs.harvard.edu/sites/default/files/research/files/harvard_jchs_short_term_era_benefits_airgood-obrycki_2022.pdf
- Bickel G, Nord M, Price C, Hamilton W, Cook J. Guide to Measuring Household Food Security (Revised 2000) . US Department of Agriculture Food and Nutrition Service; 2000. Accessed June 22, 2022. https://fns-prod.azureedge.us/sites/default/files/FSGuide.pdf
- Thorndike AN, Gardner CD, Kendrick KB, Seligman HK, Yaroch AL, Gomes AV, et al. ; American Heart Association Advocacy Coordinating Committee. Strengthening US food policies and programs to promote equity in nutrition security: a policy statement from the American Heart Association. Circulation . 2022;145(24):e1077–e1093. PubMed doi:10.1161/CIR.0000000000001072
- Drewnowski A. Food insecurity has economic root causes. Nat Food . 2022;3(8):555–556.
- Nord M, Golla AM. Does SNAP Decrease Food Insecurity? Untangling The Self-Selection Effect . Economic Research Service; 2009. Accessed March 31, 2022. https://www.ers.usda.gov/webdocs/publications/46295/10977_err85_1_.pdf?v=7939.5
- Rosenbaum D, Bergh K, Hall L. Temporary pandemic SNAP benefits will end in remaining 35 states in March 2023. Center on Budget and Policy Priorities. February 6, 2023. Accessed April 24, 2024. https://www.cbpp.org/research/food-assistance/temporary-pandemic-snap-benefits-will-end-in-remaining-35-states-in-march
- Cox R, Rodnyansky S, Henwood B, Wenzel S. Measuring population estimates of housing insecurity in the United States: a comprehensive approach. CESR-Schaeffer Working Paper; 2017. Accessed September 14, 2023. https://papers.ssrn.com/abstract=3086243
- Cornelissen S, Hermann A. COVID-19 and racial inequalities in housing: pre-pandemic and pandemic pathways to housing insecurity. J Urban Aff . 2023:1–23.
- Desmond M. Heavy is the house: rent burden among the American urban poor. Int J Urban Reg Res . 2018;42(1):160–170.
- Leopold J, Cunningham M, Posey L, Manuel T. Improving Measures of Housing Insecurity: A Path Forward . Urban Institute; 2016. Accessed January 16, 2024. https://www.urban.org/sites/default/files/publication/101608/improving_measures_of_housing_insecurity_2.pdf
- Larrimore J, Schuetz J. Assessing the severity of rent burden on low-income families. December 22, 2017. Accessed May 23, 2023. https://www.federalreserve.gov/econres/notes/feds-notes/assessing-the-severity-of-rent-burden-on-low-income-families-20171222.html
- Colburn G, Allen R. Rent burden and the Great Recession in the USA. Urban Stud . 2018;55(1):226–243.
- Shamsuddin S, Campbell C. Housing cost burden, material hardship, and well-being. Hous Policy Debate . 2022;32(3):413–432.
- Kirkpatrick SI, Tarasuk V. Housing circumstances are associated with household food access among low-income urban families. J Urban Health . 2011;88(2):284–296. PubMed doi:10.1007/s11524-010-9535-4
- Lee CY, Zhao X, Reesor-Oyer L, Cepni AB, Hernandez DC. Bidirectional relationship between food insecurity and housing instability. J Acad Nutr Diet . 2021;121(1):84–91. PubMed doi:10.1016/j.jand.2020.08.081
- King C. Food insecurity and housing instability in vulnerable families. Rev Econ Househ . 2018;16(2):255–273.
- Helms V, Coleman-Jensen A, Gray R, Brucker D. Household Food Insecurity and US Department of Housing and Urban Development Federal Housing Assistance . Economic Research Report No. ERR-277; 2011. https://scholars.unh.edu/iod/168
- Kirkpatrick SI, Tarasuk V. Adequacy of food spending is related to housing expenditures among lower-income Canadian households. Public Health Nutr . 2007;10(12):1464–1473. PubMed doi:10.1017/S136898000700081X
- Seo BK, Park GR. Food insecurity and housing affordability among low-income families: does housing assistance reduce food insecurity? Public Health Nutr . 2021;24(13):4339–4345. PubMed doi:10.1017/S1368980021001002
- Denary W, Fenelon A, Whittaker S, Esserman D, Lipska KJ, Keene DE. Rental assistance improves food security and nutrition: an analysis of national survey data. Prev Med . 2023;169:107453. PubMed doi:10.1016/j.ypmed.2023.107453
- Consolidated Appropriations Act, 2021, HR 133, 116th Congress (2019–2020). Pub L No. 116-260. Accessed April 17, 2023. https://www.congress.gov/116/plaws/publ260/PLAW-116publ260.pdf
- American Rescue Plan Act of 2021, HR 1319, 117th Congress (2021–2022), Pub L No. 117-2. Accessed April 27, 2023. https://www.congress.gov/bill/117th-congress/house-bill/1319
- Aiken C, Ellen IG, Harner I, Haupert T, Reina V, Yae R. Can emergency rental assistance be designed to prevent homelessness? Learning from emergency rental assistance programs. Hous Policy Debate . 2022;32(6):896–914.
- Aiken C, Ellen IG, Reina V. Administrative burdens in emergency rental assistance programs. RSF . 2023;9(5):100–121.
- Kneebone E, Underriner Q. An Uneven Housing Safety Net: Disparities in the Disbursement of Emergency Rental Assistance and the Role of Local Institutional Capacity . Terner Center For Housing Innovation, University of California, Berkeley; 2022. Accessed May 13, 2024. https://ternercenter.berkeley.edu/wp-content/uploads/2022/04/ERAP-April-2022-Final.pdf
- US Census Bureau. Household Pulse Survey technical documentation. Page last revised February 6, 2024. Accessed March 4, 2024. https://www.census.gov/programs-surveys/household-pulse-survey/technical-documentation.html
- US Food and Nutrition Service, US Department of Agriculture. SNAP eligibility. Page updated April 26, 2024. Accessed January 18, 2024. https://www.fns.usda.gov/snap/recipient/eligibility
- Closing the Meal Gap Act of 2023, S 1336, 118th Congress (2023–2024). Accessed April 27, 2023. https://www.congress.gov/bill/118th-congress/senate-bill/1336
- Wutich A, Brewis A. Food, water, and scarcity: toward a broader anthropology of resource insecurity. Curr Anthropol . 2014;55(4):444–468.
| Characteristic | Overall sample (N = 40,895) | Receipt of SNAP emergency allotments (EAs) | Receipt of emergency rent assistance | |||||
|---|---|---|---|---|---|---|---|---|
| SNAP participant in a state with EAs (n = 9,874) | SNAP participant in a state without EAs (n = 7,997) | Non–SNAP participant in a state with EAs (n = 11,401) | Non–SNAP participant in a state without EAs (n = 11,623) | Yes (n = 7,712) | No, waitlisted or denied (n = 3,886) | No, did not apply (n = 29,297) | ||
| Yes | 83.4 | 79.9 | 78.7 | 87.8 | 85.5 | 75.2 | 89.3 | 84.5 |
| No | 16.6 | 20.1 | 21.3 | 12.2 | 14.5 | 24.8 | 10.7 | 15.5 |
| Food sufficient | 69.2 | 69.1 | 64.6 | 70.8 | 71.0 | 61.7 | 52.2 | 73.3 |
| Food insufficient | 30.8 | 30.9 | 35.4 | 29.2 | 29.0 | 38.3 | 47.8 | 26.7 |
| Yes | 80.4 | 77.3 | 77.0 | 82.6 | 83.9 | 77.1 | 48.2 | 85.7 |
| No | 19.6 | 22.7 | 23.0 | 17.4 | 16.1 | 22.9 | 51.8 | 14.3 |
| SNAP participant in a state with EAs | 27.2 | 100.0 | — | — | — | 43.3 | 34.8 | 22.6 |
| SNAP participant in a state without EAs | 18.3 | — | 100.0 | — | — | 40.1 | 24.1 | 14.8 |
| Non–SNAP participant in a state with EAs | 30.6 | — | — | 100.0 | — | 14.3 | 23.8 | 35.3 |
| Non–SNAP participant in a state without EAs | 23.8 | — | — | — | 100.0 | 12.3 | 17.3 | 27.3 |
| Yes | 16.5 | 26.3 | 27.2 | 7.7 | 8.5 | 100.0 | — | — |
| No, waitlisted or denied | 10.3 | 13.1 | 13.5 | 8.0 | 7.5 | — | 100.0 | — |
| No, did not apply | 73.2 | 60.6 | 59.3 | 84.3 | 84.0 | — | — | 100.0 |
| ≤35 | 35.5 | 28.1 | 29.3 | 41.5 | 41.0 | 26.2 | 35.7 | 37.5 |
| 36–64 | 48.1 | 54.8 | 53.4 | 43.6 | 42.1 | 56.9 | 55.2 | 45.1 |
| ≥65 | 16.4 | 17.1 | 17.3 | 14.9 | 16.9 | 16.9 | 9.1 | 17.4 |
| Yes | 53.8 | 51.3 | 53.2 | 40.8 | 41.8 | 50.0 | 58.1 | 43.7 |
| No | 46.2 | 48.7 | 46.8 | 59.2 | 58.2 | 50.0 | 41.9 | 56.3 |
| Male | 34.2 | 27.6 | 26.3 | 41.0 | 38.9 | 26.3 | 30.3 | 36.5 |
| Female | 62.4 | 69.1 | 71.4 | 55.1 | 57.5 | 70.9 | 66.4 | 60.0 |
| Transgender or other than any listed option | 3.4 | 3.3 | 2.3 | 3.9 | 3.6 | 2.8 | 3.3 | 3.5 |
| Asian | 3.7 | 3.1 | 2.0 | 5.4 | 3.6 | 1.8 | 2.2 | 4.3 |
| Black | 23.8 | 30.0 | 31.7 | 17.1 | 19.5 | 32.7 | 36.6 | 20.1 |
| White | 63.7 | 57.2 | 57.9 | 68.3 | 69.6 | 55.7 | 48.6 | 67.6 |
| Any other single race or any combination | 8.8 | 9.7 | 8.4 | 9.2 | 7.3 | 9.8 | 12.6 | 8.0 |
| Yes | 24.2 | 23.3 | 18.9 | 29.8 | 22.3 | 20.1 | 24.9 | 25.0 |
| No | 75.8 | 76.7 | 81.1 | 70.2 | 77.7 | 79.9 | 75.1 | 75.0 |
| <25,000 | 70.6 | 79.1 | 78.1 | 64.0 | 63.4 | 79.4 | 69.2 | 68.8 |
| 25,000–34,999 | 18.4 | 14.1 | 15.0 | 21.9 | 21.7 | 13.5 | 19.4 | 19.4 |
| 35,000–49,999 | 10.2 | 6.1 | 6.2 | 13.2 | 13.9 | 6.7 | 11.1 | 10.8 |
| 50,000–74,999 | 0.8 | 0.7 | 0.7 | 0.9 | 1.0 | 0.4 | 0.3 | 1.0 |
| Yes | 21.8 | 22.7 | 22.6 | 22.2 | 19.7 | 23.1 | 38.7 | 19.1 |
| No | 78.2 | 77.3 | 77.4 | 77.8 | 80.3 | 76.9 | 61.3 | 80.9 |
| Yes | 44.3 | 30.5 | 32.0 | 56.0 | 54.7 | 32.2 | 44.0 | 47.1 |
| No | 55.7 | 69.5 | 68.0 | 44.0 | 45.3 | 67.8 | 56.0 | 52.9 |
| Less than high school or some high school | 16.8 | 21.1 | 19.0 | 15.7 | 11.5 | 18.2 | 19.4 | 16.1 |
| High school graduate or equivalent | 41.6 | 43.2 | 45.2 | 38.1 | 41.3 | 41.5 | 40.2 | 41.8 |
| Some college, but degree not received or in progress | 22.9 | 21.6 | 21.8 | 23.5 | 24.6 | 24.0 | 23.9 | 22.5 |
| Associate or bachelor’s degree | 15.4 | 12.4 | 11.9 | 17.9 | 18.4 | 14.0 | 14.2 | 15.9 |
| Graduate degree | 3.3 | 1.7 | 2.1 | 4.8 | 4.2 | 2.3 | 2.3 | 3.7 |
| Yes | 17.7 | 22.0 | 25.5 | 13.4 | 12.2 | 26.6 | 23.5 | 14.8 |
| No | 82.3 | 78.0 | 74.5 | 86.6 | 87.8 | 73.4 | 76.5 | 85.2 |
| Now married | 22.5 | 20.6 | 19.5 | 24.7 | 24.0 | 19.3 | 23.3 | 23.0 |
| Widowed | 6.4 | 6.8 | 6.4 | 5.6 | 7.1 | 7.2 | 4.9 | 6.5 |
| Divorced | 21.3 | 24.0 | 25.4 | 17.2 | 20.4 | 25.2 | 19.7 | 20.7 |
| Separated | 6.0 | 6.8 | 6.9 | 5.8 | 4.6 | 7.2 | 6.6 | 5.6 |
| Never married | 43.8 | 41.8 | 41.8 | 46.7 | 43.9 | 41.1 | 45.5 | 44.2 |
| 3.0 (1.5) | 3.1 (1.5) | 3.2 (1.7) | 3.0 (1.4) | 2.9 (1.6) | 2.9 (1.5) | 3.4 (1.5) | 3.0 (1.5) | |
Abbreviations: — , does not apply; EA, emergency allotment; SNAP, Supplemental Nutrition Assistance Program. a Low-income renters defined as renters with household incomes <130% of the federal poverty threshold. All values are percentages unless otherwise indicated. b Percentages may not sum to 100 because of rounding. c Survey question was, “Have you or anyone in your household applied for emergency rental assistance through your state or local government to cover your unpaid rent or utility bills?” d Survey question was, “During the last 7 days, did you or anyone in your household get free groceries from a food pantry, food bank, church, or other place that helps with free food?” Response options were yes or no.
| Exposure | No. of survey respondents | Predicted probability of food sufficiency, % (95% CI) | Difference in predicted probabilities, percentage point (95% CI) |
|---|---|---|---|
| Yes | 33,959 | 68.7 (68.0 to 69.5) | −3.0 (−4.8 to −1.1) |
| No | 6,936 | 71.7 (69.9 to 73.4) | Reference |
| SNAP participant in state with EAs | 9,874 | 72.4 (70.3 to 74.5) | 5.2 (1.7 to 8.7) |
| SNAP participant in state without EAs | 7,997 | 67.2 (65.2 to 69.2) | Reference |
| Non–SNAP participant in state with EAs | 11,401 | 68.8 (66.8 to 70.8) | 1.6 (−1.8 to 5.0) |
| Non–SNAP participant in state without EAs | 11,623 | 67.4 (65.2 to 69.7) | 0.2 (−1.8 to 2.3) |
| Yes | 7,712 | 64.5 (62.8 to 66.2) | 6.9 (3.9 to 9.8) |
| No, waitlisted or denied ERA | 3,886 | 57.6 (55.1 to 60.2) | Reference |
| No, did not apply | 29,297 | 72.1 (71.3 to 72.9) | 14.5 (11.8 to 17.2) |
| SNAP participant, state with EAs | 7,877 | 72.0 (69.7 to 74.3) | 5.9 (2.0 to 9.8) |
| SNAP participant, state without EAs | 6,169 | 66.0 (63.7 to 68.4) | Reference |
| Non–SNAP participant, state with EAs | 10,044 | 68.3 (66.2 to 70.5) | 2.3 (−1.5 to 6.1) |
| Non–SNAP participant, state without EAs | 9,869 | 67.6 (65.2 to 70.0) | 1.6 (−0.9 to 4.1) |
| SNAP participant, state with EAs | 1,997 | 74.0 (70.7 to 77.3) | 2.5 (−2.0 to 7.0) |
| SNAP participant, state without EAs | 1,828 | 71.5 (68.7 to 74.3) | Reference |
| Non–SNAP participant, state with EAs | 1,357 | 72.1 (67.9 to 76.3) | 0.6 (−4.3 to 5.5) |
| Non–SNAP participant, state without EAs | 1,754 | 66.8 (63.3 to 70.2) | −4.7 (−8.6 to −0.8) |
| Yes, received ERA | 5,893 | 63.0 (61.0 to 65.1) | 5.2 (2.0 to 8.5) |
| No, waitlisted or denied ERA | 3,412 | 57.8 (55.1 to 60.5) | Reference |
| No, did not apply | 24,654 | 71.7 (70.8 to 72.6) | 13.9 (10.9 to 16.9) |
| Yes, received ERA | 1,819 | 68.9 (65.3 to 72.6) | 13.0 (5.1 to 20.9) |
| No, waitlisted or denied ERA | 474 | 55.9 (49.0 to 62.8) | Reference |
| No, did not apply | 4,643 | 74.5 (72.4 to 76.6) | 18.5 (11.5 to 25.6) |
Abbreviations: EA, emergency allotment; ERA, emergency rent assistance; SNAP, Supplemental Nutrition Assistance Program. a Low-income renters defined as renters with household incomes <130% of the federal poverty threshold. Main effect and interaction models were estimated separately for each exposure/interaction; all models adjusted for demographic characteristics with fixed effects for state and survey release. b Survey question was, “Have you or anyone in your household applied for emergency rental assistance through your state or local government to cover your unpaid rent or utility bills?”
| Exposure | No. of survey respondents | Predicted probability of being current on rent, % (95% CI) | Difference in predicted probabilities, percentage point (95% CI) |
|---|---|---|---|
| Yes | 33,959 | 79.5 (78.6 to 80.4) | −5.6 (−7.3 to −3.9) |
| No | 6,936 | 85.1 (83.6 to 86.6) | Reference |
| SNAP participant, state with EAs | 9,874 | 79.6 (77.9 to 81.3) | 1.0 (−2.6 to 4.6) |
| SNAP participant, state without EAs | 7,997 | 78.6 (76.1 to 81.0) | Reference |
| Non–SNAP participant, state with EAs | 11,401 | 81.7 (80.3 to 83.2) | 3.2 (−0.2 to 6.5) |
| Non–SNAP participant, state without EAs | 11,623 | 81.4 (79.3 to 83.5) | 2.8 (0.5 to 5.2) |
| Yes, received ERA | 7,712 | 78.7 (76.9 to 80.6) | 22.4 (18.8 to 25.9) |
| No, waitlisted or denied ERA | 3,886 | 56.4 (53.5 to 59.3) | Reference |
| No, did not apply | 29,297 | 84.7 (84.0 to 85.5) | 28.3 (25.2 to 31.4) |
| SNAP participant, state with EAs | 7,877 | 78.3 (76.5 to 80.1) | 2.0 (−1.8 to 5.8) |
| SNAP participant, state without EAs | 6,169 | 76.3 (73.7 to 78.9) | Reference |
| Non–SNAP participant, state with EAs | 10,044 | 81.1 (79.6 to 82.7) | 4.8 (1.3 to 8.3) |
| Non–SNAP participant, state without EAs | 9,869 | 81.4 (79.2 to 83.5) | 5.1 (2.6 to 7.6) |
| SNAP participant, state with EAs | 1,997 | 84.5 (81.8 to 87.3) | −3.1 (−7.1 to 1.0) |
| SNAP participant, state without EAs | 1,828 | 87.6 (84.7 to 90.6) | Reference |
| Non–SNAP participant, state with EAs | 1,357 | 85.4 (82.4 to 88.5) | −2.2 (−6.9 to 2.5) |
| Non–SNAP participant, state without EAs | 1,754 | 82.7 (76.9 to 85.9) | −4.9 (−8.8 to −1.0) |
| Yes, received ERA | 5,893 | 76.7 (74.4 to 78.9) | 20.9 (16.9 to 24.8) |
| No, waitlisted/denied ERA | 3,412 | 55.8 (52.7 to 58.8) | Reference |
| No, did not apply | 24,654 | 84.1 (83.3 to 85.0) | 28.3 (25.1 to 31.5) |
| Yes, received ERA | 1,819 | 85.5 (83.3 to 87.8) | 24.8 (17.5 to 32.0) |
| No, waitlisted or denied ERA | 474 | 60.9 (53.7 to 62.8) | Reference |
| No, did not apply | 4,643 | 87.9 (86.3 to 89.5) | 27.1 (19.5 to 34.7) |
The opinions expressed by authors contributing to this journal do not necessarily reflect the opinions of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors’ affiliated institutions.
Unraveling entrepreneurial comebacks: the curvilinear relationship between entrepreneurial failure and reentry intention
- Published: 27 August 2024
Cite this article

- Shaoshuai Zhang 1 ,
- Hui Yang 1 &
- Yuan Wei ORCID: orcid.org/0009-0000-2671-5192 1
15 Accesses
Explore all metrics
Despite the powerful benefits of entrepreneurial failure experience with regard to experiential learning and future venture performance, our understanding of how failure experience impacts entrepreneurs’ decision to reenter entrepreneurship while taking advantage of the lessons that they have learned from their previous entrepreneurial endeavors remains limited. While some studies have highlighted the potential of entrepreneurial failure experience to stimulate reentry intention, other researchers have argued that failure experience can actually decrease subsequent entrepreneurial intention. This study draws on various streams of research on entrepreneurs’ responses to business failures at the cognitive, affective, and behavioral levels to propose the existence of a curvilinear relationship between entrepreneurial failure and reentry intention. We employ hierarchical regression to test a series of hypotheses by reference to a sample of 379 entrepreneurs who had experienced failure in their recent business ventures. The results reveal that the degree of failure exhibits an inverted U-shaped relationship with reentry intention. Furthermore, we find that the effect of entrepreneurial failure on reentry intention is mediated by entrepreneurs’ learning from failure and that entrepreneurial passion moderates the effects of entrepreneurial failure on both learning from failure and reentry intention. This article helps explain the distinctive effects of failure experience on reentry intention and provides empirical evidence that can facilitate the development of tailor-made support programs that can help previously failed entrepreneurs address the challenges that they encounter during the process of reentry into entrepreneurship.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Subscribe and save.
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions

Similar content being viewed by others
Managing crisis: a qualitative lens on the aftermath of entrepreneurial failure
Does entrepreneurship ecosystem influence business re-entries after failure?
Causal ascriptions and perceived learning from entrepreneurial failure, data availability.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Aguzzoli, R., Lengler, J., Sousa, C. M. P., & Benito, G. R. G. (2021). Here we go again: A case study on re-entering a foreign market. British Journal of Management , 32 (2), 416–434. https://doi.org/10.1111/1467-8551.12407
Article Google Scholar
Amankwah-Amoah, J., Boso, N., & Antwi-Agyei, I. (2018). The effects of Business failure experience on successive entrepreneurial engagements: An evolutionary phase model. Group & Organization Management , 43 (4), 648–682. https://doi.org/10.1177/1059601116643447
Amankwah-Amoah, J., Khan, Z., Ifere, S. E., Nyuur, R. B., & Khan, H. (2022). Entrepreneurs’ learning from business failures: An emerging market perspective. British Journal of Management , 33 (4), 1735–1756. https://doi.org/10.1111/1467-8551.12557
Artinger, S., & Powell, T. C. (2016). Entrepreneurial failure: Statistical and psychological explanations [Article]. Strategic Management Journal , 37 (6), 1047–1064. https://doi.org/10.1002/smj.2378
Atuahene-Gima, K., & Murray, J. Y. (2007). Exploratory and exploitative learning in new product development: A social capital perspective on new technology ventures in China. Journal of International Marketing , 15 (2), 1–29. https://doi.org/10.1509/jimk.15.2.1
Baù, M., Sieger, P., Eddleston, K. A., & Chirico, F. (2017). Fail but try again? The effects of age, gender, and multiple–owner experience on failed entrepreneurs’ reentry. Entrepreneurship Theory and Practice , 41 (6), 909–941. https://doi.org/10.1111/etap.12233
Baron, R., & Kenny, D. (1986). The moderator-mediator variable distinction in social psychological research: Conceptual, strategic, and statistical considerations. Journal of Personality and Social Psychology , 51 , 1173–1182. https://doi.org/10.1037//0022-3514.51.6.1173
Baum, J. R., & Locke, E. (2004). The relationship of entrepreneurial traits, skill, and motivation to subsequent venture growth. Journal of Applied Psychology , 89 , 587–598. https://doi.org/10.1037/0021-9010.89.4.587
Article PubMed Google Scholar
Belchior, R. F., & Castro-Silva, H. (2023). The virtuous cycle of entrepreneurial identity and experience - a longitudinal analysis. International Entrepreneurship and Management Journal , 19 (4), 1739–1770. https://doi.org/10.1007/s11365-023-00898-7
Biraglia, A., & Kadile, V. (2017). The role of entrepreneurial passion and Creativity in developing entrepreneurial intentions: Insights from American homebrewers [Article]. Journal of Small Business Management , 55 (1), 170–188. https://doi.org/10.1111/jsbm.12242
Boso, N., Adeleye, I., Donbesuur, F., & Gyensare, M. (2019). Do entrepreneurs always benefit from business failure experience? Journal of Business Research , 98 , 370–379. https://doi.org/10.1016/j.jbusres.2018.01.063
Brüderl, J., Preisendörfer, P., & Ziegler, R. (1992). Survival chances of newly founded business organizations. American Sociological Review , 57 (2), 227–242. https://doi.org/10.2307/2096207
Byrne, O., & Shepherd, D. A. (2015). Different strokes for different folks: Entrepreneurial narratives of emotion, cognition, and making sense of business failure. Entrepreneurship Theory and Practice , 39 (2), 375–405. https://doi.org/10.1111/etap.12046
Cardon, M. S., Gregoire, D. A., Stevens, C. E., & Patel, P. C. (2013). Measuring entrepreneurial passion: Conceptual foundations and scale validation. Journal of Business Venturing , 28 (3), 373–396. https://doi.org/10.1016/j.jbusvent.2012.03.003
Cardon, M. S., & Kirk, C. P. (2015). Entrepreneurial passion as mediator of the self-efficacy to persistence relationship. Entrepreneurship: Theory and Practice , 39 (5), 1027–1050. https://doi.org/10.1111/etap.12089
Cardon, M. S., Stevens, C. E., & Potter, D. R. (2011). Misfortunes or mistakes? Cultural sensemaking of entrepreneurial failure. Journal of Business Venturing , 26 (1), 79–92. https://doi.org/10.1016/j.jbusvent.2009.06.004
Cardon, M. S., Wincent, J., Singh, J., & Drnovsek, M. (2009). The nature and experience of entrepreneurial passion. Academy of Management Review , 34 (3), 511–532. https://doi.org/10.5465/AMR.2009.40633190
Chen, C. C., Greene, P. G., & Crick, A. (1998). Does entrepreneurial self-efficacy distinguish entrepreneurs from managers? [Article]. Journal of Business Venturing , 13 (4), 295–316. https://doi.org/10.1016/S0883-9026(97)00029-3
Cope, J. (2011). Entrepreneurial learning from failure: An interpretative phenomenological analysis. Journal of Business Venturing , 26 (6), 604–623. https://doi.org/10.1016/j.jbusvent.2010.06.002
Corner, P. D., Singh, S., & Pavlovich, K. (2017). Entrepreneurial resilience and venture failure. International Small Business Journal-Researching Entrepreneurship , 35 (6), 687–708. https://doi.org/10.1177/0266242616685604
Costa, P. L., Ferreira, J. J., & de Torres, R. (2023). From entrepreneurial failure to re-entry [Article]. Journal of Business Research , 158 . https://doi.org/10.1016/j.jbusres.2023.113699 . Article 113699.
De Hoe, R., & Janssen, F. (2022). Re-creation after business failure: A conceptual model of the mediating role of psychological capital. Frontiers in Psychology , 13 , 842590. https://doi.org/10.3389/fpsyg.2022.842590
De Sordi, J. O., dos Santos, A. R., de Azevedo, M. C., Jorge, C. F. B., & Hashimoto, M. (2022). Dark, down, and destructive side of entrepreneurship: Unveiling negative aspects of unsuccessful entrepreneurial action. International Journal of Management Education , 20 (3), 100659. https://doi.org/10.1016/j.ijme.2022.100659
Drnovsek, M., Cardon, M. S., & Patel, P. C. (2016). Direct and indirect effects of passion on growing technology ventures. Strategic Entrepreneurship Journal , 10 (2), 194–213. https://doi.org/10.1002/sej.1213
Edeh, F. O., Zayed, N. M., Darwish, S., Nitsenko, V., Hanechko, I., & Islam, K. M. A. (2023). Impression management and employee contextual performance in service organizations (enterprises). Emerging Science Journal , 7 (2), 366–384. https://doi.org/10.28991/esj-2023-07-02-05
Eftekhari, N., & Timmermans, B. (2022). New venture dissolution and the comobility of new venture teams. Small Business Economics , 59 (1), 279–298. https://doi.org/10.1007/s11187-021-00543-z
Eggers, J. P., & Song, L. (2015). Dealing with failure: Serial entrepreneurs and the costs of changing industries between ventures [Article]. Academy of Management Journal , 58 (6), 1785–1803. https://doi.org/10.5465/amj.2014.0050
Espinoza-Benavides, J., & Díaz, D. (2019). The entrepreneurial profile after failure. International Journal of Entrepreneurial Behavior & Research , 25 (8), 1634–1651. https://doi.org/10.1108/ijebr-04-2018-0242
Fanaja, R. A., Pradana, M., Eka Saputri, M., & Utami, D. G. (2023). Knowledge management as driver of women’s entrepreneurial innovativeness. Journal of Human Earth and Future , 4 (1), 1–9. https://doi.org/10.28991/hef-2023-04-01-01
Fan-Osuala, O. (2021). All failures are not equal: Degree of failure and the launch of subsequent crowdfunding campaigns [Article]. Journal of Business Venturing Insights , 16 . https://doi.org/10.1016/j.jbvi.2021.e00260 . Article e00260.
Fisher, R., Merlot, E., & Johnson, L. W. (2018). The obsessive and harmonious nature of entrepreneurial passion. International Journal of Entrepreneurial Behavior & Research , 24 (1), 22–40. https://doi.org/10.1108/IJEBR-01-2017-0011
Franco, M., Haase, H., & António, D. (2021). Influence of failure factors on entrepreneurial resilience in Angolan micro, small and medium-sized enterprises [Article]. International Journal of Organizational Analysis , 29 (1), 240–259. https://doi.org/10.1108/IJOA-07-2019-1829
Fu, H., Xiao, X. H., Ye, B. H. B., Fang, S. J., Li, Y. Q., & Wu, Y. Y. (2023). Stay passionate and carry on: Why passion exhausts and how it can be restored. Current Psychology , 42 (31), 27574–27592. https://doi.org/10.1007/s12144-022-03889-z
Guerrero, M., & Peña-Legazkue, I. (2019). Renascence after post-mortem: The choice of accelerated repeat entrepreneurship [Article]. Small Business Economics , 52 (1), 47–65. https://doi.org/10.1007/s11187-018-0015-7
Haans, R., Pieters, C., & He, Z. L. (2016). Thinking about U: Theorizing and Testing U- and inverted U-shaped relationships in strategy research. Strategic Management Journal , 37 , 1177–1195. https://doi.org/10.1002/smj.2399
Henriquez-Daza, M. C., Capelleras, J. L., & Osorio-Tinoco, F. (2023). Does fear of failure affect entrepreneurial growth aspirations? The moderating role of institutional collectivism in emerging and developed countries [Article]. Journal of Entrepreneurship in Emerging Economies . https://doi.org/10.1108/JEEE-08-2022-0232
He, V. F., Sirén, C., Singh, S., Solomon, G., & von Krogh, G. (2018). Keep calm and carry on: Emotion regulation in entrepreneurs’ learning from failure [Article]. Entrepreneurship: Theory and Practice , 42 (4), 605–630. https://doi.org/10.1111/etap.12273
Hsu, D. K., Shinnar, R. S., & Anderson, S. E. (2019). I wish I had a regular job’: An exploratory study of entrepreneurial regret [Article]. Journal of Business Research , 96 , 217–227. https://doi.org/10.1016/j.jbusres.2018.11.006
Hsu, D. K., Wiklund, J., & Cotton, R. D. (2017). Success, failure, and entrepreneurial reentry: An experimental assessment of the veracity of self-efficacy and prospect theory [Article]. Entrepreneurship: Theory and Practice , 41 (1), 19–47. https://doi.org/10.1111/etap.12166
Hurmerinta, L., Nummela, N., & Paavilainen-Mäntymäki, E. (2024). Boosted by failure? Entrepreneurial internationalisation as a cyclical learning process. European Journal of International Management , 22 (3). https://doi.org/10.1504/ejim.2024.136483
Hwang, K., & Choi, J. (2021). How do failed entrepreneurs cope with their prior failure when they seek subsequent re-entry into serial entrepreneurship? Failed entrepreneurs’ optimism and defensive pessimism and coping humor as a moderator. International Journal of Environmental Research and Public Health , 18 (13), 7021. https://doi.org/10.3390/ijerph18137021
Jenkins, A., & McKelvie, A. (2016). What is entrepreneurial failure? Implications for future research. International Small Business Journal-Researching Entrepreneurship , 34 (2), 176–188. https://doi.org/10.1177/0266242615574011
Jenkins, A. S., Wiklund, J., & Brundin, E. (2014). Individual responses to firm failure: Appraisals, grief, and the influence of prior failure experience. Journal of Business Venturing , 29 (1), 17–33. https://doi.org/10.1016/j.jbusvent.2012.10.006
Karimi, S. (2020). The role of entrepreneurial passion in the formation of students’ entrepreneurial intentions. Applied Economics , 52 (3), 331–344. https://doi.org/10.1080/00036846.2019.1645287
Kawai, N., Sibunruang, H., & Kazumi, T. (2023). Work-family conflict, entrepreneurial regret, and entrepreneurial outcomes during the COVID-19 pandemic [Article]. International Entrepreneurship and Management Journal , 19 (2), 837–861. https://doi.org/10.1007/s11365-023-00846-5
Article PubMed Central Google Scholar
Kiani, A., Ali, A., Biraglia, A., & Wang, D. (2023). Why I persist while others leave? Investigating the path from passion to persistence in entrepreneurship. Journal of Small Business Management , 61 (6), 2818–2848. https://doi.org/10.1080/00472778.2021.1938097
Lafuente, E., Vaillant, Y., Vendrell-Herrero, F., & Gomes, E. (2019). Bouncing back from failure: Entrepreneurial resilience and the internationalisation of subsequent ventures created by serial entrepreneurs. Applied Psychology , 68 (4), 658–694. https://doi.org/10.1111/apps.12175
Lattacher, W., & Wdowiak, M. A. (2020). Entrepreneurial learning from failure. A systematic review. International Journal of Entrepreneurial Behavior & Research , 26 (5), 1093–1131. https://doi.org/10.1108/IJEBR-02-2019-0085
Lee, C. K., Cottle, G. W., Simmons, S. A., & Wiklund, J. (2021). Fear not, want not: Untangling the effects of social cost of failure on high-growth entrepreneurship [Article]. Small Business Economics , 57 (1), 531–553. https://doi.org/10.1007/s11187-020-00324-0
Lin, S., & Wang, S. (2019). How does the age of serial entrepreneurs influence their re-venture speed after a business failure? [Article]. Small Business Economics , 52 (3), 651–666. https://doi.org/10.1007/s11187-017-9977-0
Liu, Y., Li, Y., Hao, X., & Zhang, Y. (2019). Narcissism and learning from entrepreneurial failure [Article]. Journal of Business Venturing , 34 (3), 496–512. https://doi.org/10.1016/j.jbusvent.2019.01.003
Mandl, C., Berger, E. S. C., & Kuckertz, A. (2016). Do you plead guilty? Exploring entrepreneurs’ sensemaking-behavior link after business failure [Article]. Journal of Business Venturing Insights , 5 , 9–13. https://doi.org/10.1016/j.jbvi.2015.12.002
Mantere, S., Aula, P., Schildt, H., & Vaara, E. (2013). Narrative attributions of entrepreneurial failure. Journal of Business Venturing , 28 (4), 459–473. https://doi.org/10.1016/j.jbusvent.2012.12.001
McGrath, R. G. (1999). Falling forward: Real options reasoning and entrepreneurial failure. Academy of Management Review , 24 (1), 13–30. https://doi.org/10.5465/amr.1999.1580438
Mueller, B. A., & Shepherd, D. A. (2016). Making the most of failure experiences: Exploring the relationship between business failure and the identification of business opportunities. Entrepreneurship Theory and Practice , 40 (3), 457–487. https://doi.org/10.1111/etap.12116
Mueller, B. A., Wolfe, M. T., & Syed, I. (2017). Passion and grit: An exploration of the pathways leading to venture success. Journal of Business Venturing , 32 (3), 260–279. https://doi.org/10.1016/j.jbusvent.2017.02.001
Murnieks, C. Y., Mosakowski, E., & Cardon, M. S. (2014). Pathways of passion: Identity centrality, passion, and behavior among entrepreneurs. Journal of Management , 40 (6), 1583–1606. https://doi.org/10.1177/0149206311433855
Newman, A., Obschonka, M., Moeller, J., & Chandan, G. G. (2021). Entrepreneurial passion: A review, synthesis, and agenda for future research. Applied Psychology , 70 (2), 816–860. https://doi.org/10.1111/apps.12236
Pimentel, L., Major, M., & Cruz, A. (2023). Collective action in institutional entrepreneurship: The case of a government agency. Emerging Science Journal , 7 (2), 538–557. https://doi.org/10.28991/esj-2023-07-02-017
Plehn-Dujowich, J. (2010). A theory of serial entrepreneurship. Small Business Economics , 35 (4), 377–398. https://doi.org/10.1007/s11187-008-9171-5
Politis, D. (2005). The process of entrepreneurial learning: A conceptual framework. Entrepreneurship Theory and Practice , 29 (4), 399–424. https://doi.org/10.1111/j.1540-6520.2005.00091.x
Politis, D., & Gabrielsson, J. (2009). Entrepreneurs’ attitudes towards failure - an experiential learning approach. International Journal of Entrepreneurial Behaviour & Research , 15 (4), 364–383. https://doi.org/10.1108/13552550910967921
Rawal, A., Sarpong, D., & Singh, S. K. (2023). Phoenix rising: Rebounding to venture again post firm-failure. Industrial Marketing Management , 112 , 71–84. https://doi.org/10.1016/j.indmarman.2023.05.007
Sarasvathy, S. D., Menon, A. R., & Kuechle, G. (2013). Failing firms and successful entrepreneurs: Serial entrepreneurship as a temporal portfolio [Article]. Small Business Economics , 40 (2), 417–434. https://doi.org/10.1007/s11187-011-9412-x
Saylors, R., Lahiri, A., Warnick, B., & Baid, C. (2023). Looking back to venture forward: Exploring idea and identity work in public failure narratives. Entrepreneurship Theory and Practice , 47 (2), 398–429. https://doi.org/10.1177/10422587211057027 . Article 10422587211057027.
Seckler, C., Funken, R., & Gielnik, M. (2017). Learning from entrepreneurial failure: Integrating emotional, motivational, and cognitive factors. In J. E. Ellingson & R. A. Noe (Eds.), Autonomous learning in the workplace (pp. 54–77). Routledge. https://doi.org/10.4324/9781315674131-4
Shahid, S., & Kundi, Y. M. (2022). Feel dragged out: A recovery perspective in the relationship between emotional exhaustion and entrepreneurial exit [Article]. Journal of Small Business and Enterprise Development , 29 (2), 203–220. https://doi.org/10.1108/JSBED-05-2021-0199
Shepherd, D. A. (2003). Learning from business failure: Propositions of grief recovery for the self-employed. Academy of Management Review , 28 (2), 318–328. https://doi.org/10.5465/amr.2003.9416377
Shepherd, D. A., & Patzelt, H. (2015). Harsh evaluations of entrepreneurs who fail: The role of sexual orientation, use of environmentally friendly technologies, and observers’ perspective taking. Journal of Management Studies , 52 (2), 253–284. https://doi.org/10.1111/joms.12103
Shepherd, D. A., Wiklund, J., & Haynie, J. M. (2009). Moving forward: Balancing the financial and emotional costs of business failure [Article]. Journal of Business Venturing , 24 (2), 134–148. https://doi.org/10.1016/j.jbusvent.2007.10.002
Shirshitskaia, E., Zhou, X., & Zhang, L. (2021). The impact of learning from failure on new ventures’ sustainable development. Frontiers in Psychology , 12 . https://doi.org/10.3389/fpsyg.2021.784518 . Original Research.
Shore, A., Pittaway, L., & Bortolotti, T. (2023). From negative emotions to personal growth: Failure and re-entry into Entrepreneurship. British Journal of Management . https://doi.org/10.1111/1467-8551.12785
Simmons, S. A., Wiklund, J., & Levie, J. (2014). Stigma and business failure: Implications for entrepreneurs’ career choices. Small Business Economics , 42 (3), 485–505. https://doi.org/10.1007/s11187-013-9519-3
Simmons, S. A., Wiklund, J., Levie, J., Bradley, S. W., & Sunny, S. A. (2019). Gender gaps and reentry into entrepreneurial ecosystems after business failure [Article]. Small Business Economics , 53 (2), 517–531. https://doi.org/10.1007/s11187-018-9998-3
Singh, S., Corner, P. D., & Pavlovich, K. (2015). Failed, not finished: A narrative approach to understanding venture failure stigmatization. Journal of Business Venturing , 30 (1), 150–166. https://doi.org/10.1016/j.jbusvent.2014.07.005
Singh, S., Corner, P., & Pavlovich, K. (2007). Coping with entrepreneurial failure. Journal of Management & Organization , 13 (4), 331–344. https://doi.org/10.5172/jmo.2007.13.4.331
Slade Shantz, A., Zietsma, C., Kistruck, G. M., & Cruz, L. B. (2024). Exploring the relative efficacy of ‘within-logic contrasting’ and ‘cross-logic analogizing’ framing tactics for adopting new entrepreneurial practices in contexts of poverty. Journal of Business Venturing , 39 (1). https://doi.org/10.1016/j.jbusvent.2023.106341
Smollan, R., & Singh, S. (2024). How social entrepreneurs respond to enterprise failure [Article]. Journal of Social Entrepreneurship , 15 (1), 1–25. https://doi.org/10.1080/19420676.2021.1890189
Souitaris, V., Zerbinati, S., & Al-Laham, A. (2007). Do entrepreneurship programmes raise entrepreneurial intention of science and engineering students? The effect of learning, inspiration and resources. Journal of Business Venturing , 22 (4), 566–591. https://doi.org/10.1016/j.jbusvent.2006.05.002
Stroe, S., Sirén, C., Shepherd, D., & Wincent, J. (2020). The dualistic regulatory effect of passion on the relationship between fear of failure and negative affect: Insights from facial expression analysis. Journal of Business Venturing , 35 (4), 105948. https://doi.org/10.1016/j.jbusvent.2019.105948
Tu, Y., Hao, X., Rosak-Szyrocka, J., Vasa, L., & Zhao, X. (2023). Obsessive passion, opportunity recognition, and entrepreneurial performance: The dual moderating effect of the fear of failure [Article]. Frontiers in Psychology , 13 , 1037250. https://doi.org/10.3389/fpsyg.2022.1037250
Ucbasaran, D., Shepherd, D. A., Lockett, A., & Lyon, S. J. (2013). Life after business failure: The process and consequences of business failure for entrepreneurs. Journal of Management , 39 (1), 163–202. https://doi.org/10.1177/0149206312457823
Ucbasaran, D., Westhead, P., & Wright, M. (2009). The extent and nature of opportunity identification by experienced entrepreneurs [Article]. Journal of Business Venturing , 24 (2), 99–115. https://doi.org/10.1016/j.jbusvent.2008.01.008
Ucbasaran, D., Westhead, P., Wright, M., & Flores, M. (2010). The nature of entrepreneurial experience, business failure and comparative optimism [Article]. Journal of Business Venturing , 25 (6), 541–555. https://doi.org/10.1016/j.jbusvent.2009.04.001
Vaillant, Y., Mora-Esquivel, R., & Alvarado, M. (2024). The forgetting curve in entrepreneurship: Decaying learning benefits of past entrepreneurial experience. Small Business Economics . https://doi.org/10.1007/s11187-024-00890-7
Walsh, G. S., & Cunningham, J. A. (2017). Regenerative failure and attribution: Examining the underlying processes affecting entrepreneurial learning. International Journal of Entrepreneurial Behavior & Research , 23 (4), 688–707. https://doi.org/10.1108/ijebr-03-2015-0072
Walsh, G. S., & Cunningham, J. A. (2023). Business failure and entrepreneur experiences of passion. International Small Business Journal-Researching Entrepreneurship . https://doi.org/10.1177/02662426231194482
Walter, S. G., & Block, J. H. (2016). Outcomes of entrepreneurship education: An institutional perspective. Journal of Business Venturing , 31 (2), 216–233. https://doi.org/10.1016/j.jbusvent.2015.10.003
Wang, H., Zheng, C., Wu, W., & Sui, F. (2022). How entrepreneurs’ dual narcissism affects new venture growth: The roles of personal initiative and learning from entrepreneurial failure [Article]. Journal of Organizational Change Management , 35 (7), 1125–1146. https://doi.org/10.1108/JOCM-10-2021-0313
Williams, T. A., Thorgren, S., & Lindh, I. (2020). Rising from failure, staying down, or more of the same? An inductive study of entrepreneurial reentry. Academy of Management Discoveries , 6 (4), 631–662. https://doi.org/10.5465/amd.2018.0047
Yamakawa, Y., & Cardon, M. S. (2015). Causal ascriptions and perceived learning from entrepreneurial failure. Small Business Economics , 44 (4), 797–820. https://doi.org/10.1007/s11187-014-9623-z
Yamakawa, Y., Peng, M. W., & Deeds, D. L. (2015). Rising from the ashes: Cognitive determinants of venture growth after entrepreneurial failure [Article]. Entrepreneurship: Theory and Practice , 39 (2), 209–236. https://doi.org/10.1111/etap.12047
Yang, T., & Aldrich, H. E. (2012). Out of sight but not out of mind: Why failure to account for left truncation biases research on failure rates [Article]. Journal of Business Venturing , 27 (4), 477–492. https://doi.org/10.1016/j.jbusvent.2012.01.001
Yao, K., Li, X., & Liang, B. (2021). Failure learning and entrepreneurial resilience: The moderating role of firms’ knowledge breadth and knowledge depth [Article]. Journal of Knowledge Management , 25 (9), 2141–2160. https://doi.org/10.1108/JKM-10-2020-0772
Zhao, H., & Wibowo, A. (2021). Entrepreneurship resilience: Can psychological traits of entrepreneurial intention support overcoming entrepreneurial failure? Frontiers in Psychology , 12 , Article 707803. https://doi.org/10.3389/fpsyg.2021.707803
Download references
Acknowledgements
The first author would like to acknowledge the financial support of the National Natural Science Foundation of China (Grant No: 72172165) and the Natural Science Foundation of Guangdong Province, China (Grant No: 2024A1515011283).
Author information
Authors and affiliations.
School of Business, Sun Yat-sen University, No.135 Xingang West Road, Guangzhou, 510275, Guangdong, China
Hui Fu, Min Xu, Shaoshuai Zhang, Hui Yang & Yuan Wei
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Yuan Wei .
Ethics declarations
Ethical approval.
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
We’ve obtained inform consent from all individual participants included in the study.
Conflict of interest
There is no conflict of interest.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Fu, H., Xu, M., Zhang, S. et al. Unraveling entrepreneurial comebacks: the curvilinear relationship between entrepreneurial failure and reentry intention. Curr Psychol (2024). https://doi.org/10.1007/s12144-024-06511-6
Download citation
Accepted : 31 July 2024
Published : 27 August 2024
DOI : https://doi.org/10.1007/s12144-024-06511-6
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Entrepreneurial failure
- Reentry intention
- Learning from failure
- Entrepreneurial passion
- Find a journal
- Publish with us
- Track your research

IMAGES
VIDEO
COMMENTS
2. RESEARCH DESIGN. This research is exploratory in nature as it attempts to explore the experiences of mothers of incest survivors. Their subjective perceptions formed the core data of the study; hence it needed the method that would deal with the topic in an exploratory nature. For the purpose of this study, the research paradigm that was ...
Research methodology formats can vary depending on the specific requirements of the research project, but the following is a basic example of a structure for a research methodology section: I. Introduction. Provide an overview of the research problem and the need for a research methodology section; Outline the main research questions and ...
Table of contents. Step 1: Introduce your topic. Step 2: Describe the background. Step 3: Establish your research problem. Step 4: Specify your objective (s) Step 5: Map out your paper. Research paper introduction examples. Frequently asked questions about the research paper introduction.
Step 1: Explain your methodological approach. Step 2: Describe your data collection methods. Step 3: Describe your analysis method. Step 4: Evaluate and justify the methodological choices you made. Tips for writing a strong methodology chapter. Other interesting articles.
Research Methodology Example. Detailed Walkthrough + Free Methodology Chapter Template. If you're working on a dissertation or thesis and are looking for an example of a research methodology chapter, you've come to the right place. In this video, we walk you through a research methodology from a dissertation that earned full distinction ...
The methodologies will include areas such as the location of the study, research design, sampling and sample size, types of data, data collection method and its management. 3.1 Research Design ...
Do yourself a favour and start with the end in mind. Section 1 - Introduction. As with all chapters in your dissertation or thesis, the methodology chapter should have a brief introduction. In this section, you should remind your readers what the focus of your study is, especially the research aims. As we've discussed many times on the blog ...
1.2 Defining Research Methodology. It is an essential process of any scientific study, which serves as a framework for processing and achieving the predicted outcomes of the study. It is commonly defined as a systematic and organized process of collecting, analyzing, interpreting, and presenting information to answer specific questions or solve ...
The research design is a fundamental aspect of research methodology, outlining the overall strategy and structure of the study. It includes decisions regarding the research type (e.g., descriptive, experimental), the selection of variables, and the determination of the study's scope and timeframe. We must carefully consider the design to ...
Research methods are specific procedures for collecting and analyzing data. Developing your research methods is an integral part of your research design. When planning your methods, there are two key decisions you will make. First, decide how you will collect data. Your methods depend on what type of data you need to answer your research question:
As we mentioned, research methodology refers to the collection of practical decisions regarding what data you'll collect, from who, how you'll collect it and how you'll analyse it. Research design, on the other hand, is more about the overall strategy you'll adopt in your study. For example, whether you'll use an experimental design ...
Research methodology is a systematic investigation to find ways to a problem (Burns, 2000). There are many possible ways to look at the research methodology. However the ... 3.7.2.A Sample Selection 3-9. Chapter 3 Research Methodology In general, potential members of a CoP go through a self-selection process, assessing ...
2.1 Research Methodology. Method can be described as a set of tools and techniques for finding something out, or for reducing levels of uncertainty. According to Saunders (2012) method is the technique and procedures used to obtain and analyse research data, including for example questionnaires, observation, interviews, and statistical and non-statistical techniques [].
3.1 Introduction. This chapter presents the methodology which was employed during the study. In. light of this, the areas of the study and reasons which underpin the choice of area. are explained ...
Definition, Types, and Examples. Research methodology 1,2 is a structured and scientific approach used to collect, analyze, and interpret quantitative or qualitative data to answer research questions or test hypotheses. A research methodology is like a plan for carrying out research and helps keep researchers on track by limiting the scope of ...
METHODOLOGY 3.1 Introduction Research can be defined as the systematic process of collecting and logically analysing data for ... sample, research instruments and methods of data collection utilised in the study. This chapter also provides a description of how the data will be analysed, and discusses the issue of reliability and validity as
In other words, the research design sets the procedure on the required data, the methods to be applied to collect and analyze this data, and how all of this is going to answer the research ...
Chapter 2 Introduction. Chapter 2. Introduction. Maybe you have already gained some experience in doing research, for example in your bachelor studies, or as part of your work. The challenge in conducting academic research at masters level, is that it is multi-faceted. The types of activities are: Writing up and presenting your findings.
3. CHAPTER 3. RCH METHODOLOGY3.1 IntroductionThis Chapter presents the de. It provides. d in undertaking this research aswell as a justifi. on for the use of this method. lection of participants, the datacollection process. nd the process of data analysi. . The Chapter also discusses therole of the researcher in qualitative re.
Introduction Research methodology is the indispensable part of any research work. This guides the researcher about the flow of research and provides the framework through which the research is to be carried out. This chapter expounds the research paradigm, research approach, research design, ... •Sample Size: 30 Qualitative; 400 Quantitative
research methods. Since the object of research, particularly the applied research, it to arrive at a solution for a given problem, the available data and the unknown aspects of the problem have to be related to each other to make a solution possible. Keeping this in view, research methods can be put into the following three groups: 1.
View PDF. CHAPTER THREE RESEARCH METHODOLOGY Introduction This chapter presents the methodology used for collecting the data for the study. It explains the study population, sample and sampling procedure, data collection techniques, data collection procedure, administration of the survey instrument, and data analysis.
People are described using language that affirms their worth and dignity. Authors plan for ethical compliance and report critical details of their research protocol to allow readers to evaluate findings and other researchers to potentially replicate the studies. Tables and figures present information in an engaging, readable manner.
Bovine leukemia virus (BLV) is the etiological agent of enzootic bovine leukosis and causes a persistent infection that can leave cattle with no symptoms. Many countries have been able to successfully eradicate BLV through improved detection and management methods. However, with the increasing novel molecular detection methods there have been few efforts to standardize these results at global ...
A research methodology is defined as the study of how scientific research is conducted. According to Mishra and Alok (2022), a research methodology outlines what research is about, how to proceed ...
Recent advancements in translational gut microbiome research have revealed its crucial role in shaping predictive healthcare applications. Herein, we introduce the Gut Microbiome Wellness Index 2 ...
The sensory evaluation followed the Methodology for Sensory Evaluation of Tea in the National Standard of the People's Republic of China (GB/T 23776-2018). Each tea sample was weighed to 3 g and steeped in 150 ml evaluation cups filled with boiling water at 100°C for 5 minutes before being ready for evaluation. 2.3. Chemicals
Methods. We used data from the US Household Pulse Survey (June 2022-May 2023) to examine whether associations between emergency support policies and indicators of food and housing security differed according to household rent burden status. ... Sample. Our sample consisted of renters with incomes less than 130% of the federal poverty ...
In this work, we generate a single attosecond electron pulse by the optical gating approach using a polarization-gated half-cycle laser pulse (31, 44, 45).Then, we demonstrate the attained attosecond temporal resolution by measuring the attosecond electron diffraction to probe the sub-half-cycle field-driven electron dynamics of multilayer single crystalline graphene (fig. S1).
Research procedure and sample. As the focus of our research is on the impact of entrepreneurial failure on reentry intention, in line with previous studies (Shore et al., 2023), a cross-sectional survey was conducted to collect sensitive data regarding entrepreneurial failure experiences from entrepreneurs.To collect such data, questionnaires were sent to target respondents (i.e ...